













| General Information: |
|
| Name(s) found: |
cbh1 /
SPAC9E9.10c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 514 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA condensed nuclear chromosome, centromeric region [IDA nuclear chromatin [NAS spindle pole body [IDA spindle [IDA |
| Biological Process: |
chromatin silencing at centromere
[IMP histone modification [IMP mitotic sister chromatid segregation [IGI |
| Molecular Function: |
protein binding
[IPI protein dimerization activity [NAS centromeric DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPIRQAVSL AEKKAIRDYY NQSAHRIPQK EVTEWFRKTY NKDLSQSTIS EILSPKYSYL 60
61 DDGSVRLGDI KKIRAPKFPL LDNAVYEWLQ QREVEGLPIS GDMIKQAATR FWSKIPAYAS 120
121 LPLPDFSNGW LDKFRRRHYI QQSAVNQALE SIKFEVFREY PVHIQEFIRP YAQKDIFCVG 180
181 GTSLFWKLTP NKQNLDYQEI QGMKRGKAKV SLIMCCNADG SERLPLWIVG YAQNPRSFEQ 240
241 CGIFPKSMNF EWKWNGRASI SPIIMEEWLR WFDTRMQGRK VLLMLESNDT YQFGVESVRR 300
301 FKGGFQNTSV FRIPEKTLDI KSPFEQGIVD TFKANYRRYW LQYSLNQINI LRDPLKAVNV 360
361 LKAIRWMLWS WNFGVSEKTI QASFLRSGLL GPHSEAEGQG MESYQKAIME IGNLISSKGS 420
421 EAVKQINEYI RPSEEDETKL HQDAVDTVAA EFLEERRFES DEEEDVEPQI SNVEAAMAFE 480
481 VLIKYFEQHE NGDPSFVLDL YRRQRVIAPN NLIA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
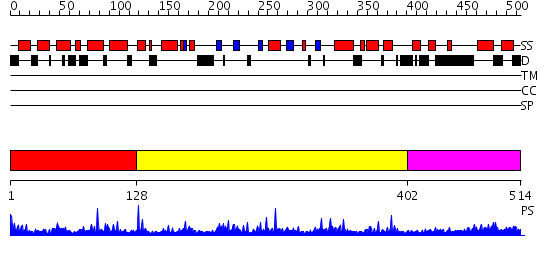
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | 24.69897 | Ars-binding protein 1, ABP1 | |
| 2 | View Details | [128..401] | 1.43 | No description for 2f7tA was found. | |
| 3 | View Details | [402..514] | 3.150996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)