













| General Information: |
|
| Name(s) found: |
shi-PK /
FBpp0290810
[FlyBase]
shi-PJ / FBpp0290809 [FlyBase] shi-PF / FBpp0089280 [FlyBase] shi-PG / FBpp0089277 [FlyBase] |
| Description(s) found:
Found 52 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 877 amino acids |
Gene Ontology: |
|
| Cellular Component: |
sperm individualization complex
[IDA]
synapse [IMP] microtubule associated complex [ISS] plasma membrane [IDA] |
| Biological Process: |
sperm individualization
[IGI][IMP]
male courtship behavior [IMP] pole cell formation [TAS] cytokinesis [IMP][TAS] border follicle cell migration [IMP] memory [IMP][TAS] microtubule-based process [NAS] learning or memory [IDA] salivary gland morphogenesis [IMP] epithelial cell migration, open tracheal system [IMP] response to heat [IMP] male courtship behavior, veined wing extension [IMP] vesicle-mediated transport [TAS] synaptic vesicle endocytosis [IMP][TAS] olfactory learning [IDA] male courtship behavior, veined wing generated song production [IMP] endocytosis [IMP][NAS][TAS] cellularization [IMP][TAS] receptor-mediated endocytosis [NAS] conditioned taste aversion [IMP] open tracheal system development [IMP] larval feeding behavior [IMP] short-term memory [IMP] regulation of actin cytoskeleton organization [IGI] proboscis extension reflex [IMP] synaptic vesicle membrane organization [IMP] synaptic vesicle budding from presynaptic membrane [IMP][TAS] regulation of synapse structure and activity [IMP] extracellular matrix organization [IMP] morphogenesis of a polarized epithelium [IMP] adherens junction maintenance [IMP] regulation of actin filament-based process [IGI] regulation of short-term neuronal synaptic plasticity [IMP] |
| Molecular Function: |
GTP binding
[IEA]
microtubule motor activity [NAS] actin binding [IDA] GTPase activity [ISS][NAS] microtubule binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSLITIVNK LQDAFTSLGV HMQLDLPQIA VVGGQSAGKS SVLENFVGKD FLPRGSGIVT 60
61 RRPLILQLIN GVTEYGEFLH IKGKKFSSFD EIRKEIEDET DRVTGSNKGI SNIPINLRVY 120
121 SPHVLNLTLI DLPGLTKVAI GDQPVDIEQQ IKQMIFQFIR KETCLILAVT PANTDLANSD 180
181 ALKLAKEVDP QGVRTIGVIT KLDLMDEGTD ARDILENKLL PLRRGYIGVV NRSQKDIEGR 240
241 KDIHQALAAE RKFFLSHPSY RHMADRLGTP YLQRVLNQQL TNHIRDTLPG LRDKLQKQML 300
301 TLEKEVEEFK HFQPGDASIK TKAMLQMIQQ LQSDFERTIE GSGSALVNTN ELSGGAKINR 360
361 IFHERLRFEI VKMACDEKEL RREISFAIRN IHGIRVGLFT PDMAFEAIVK RQIALLKEPV 420
421 IKCVDLVVQE LSVVVRMCTA KMSRYPRLRE ETERIITTHV RQREHSCKEQ ILLLIDFELA 480
481 YMNTNHEDFI GFANAQNKSE NANKTGTRQL GNQVIRKGHM VIQNLGIMKG GSRPYWFVLT 540
541 SESISWYKDE DEKEKKFMLP LDGLKLRDIE QGFMSMSRRV TFALFSPDGR NVYKDYKQLE 600
601 LSCETVEDVE SWKASFLRAG VYPEKQETQE NGDESASEES SSDPQLERQV ETIRNLVDSY 660
661 MKIVTKTTRD MVPKAIMMLI INNAKDFING ELLAHLYASG DQAQMMEESA ESATRREEML 720
721 RMYRACKDAL QIIGDVSMAT VSSPLPPPVK NDWLPSGLDN PRLSPPSPGG VRGKPGPPAQ 780
781 SSLGGRNPPL PPSTGRPAPA IPNRPGGGAP PLPGGRPGGS LPPPMLPSRV SGAVGGAIVQ 840
841 QSGANRYVPE SMRGQVNQAV GQAAINELSN AFSSRFK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
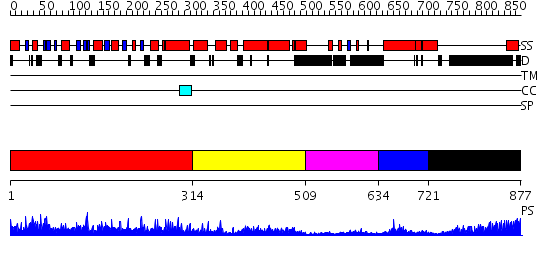
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..313] | 131.0 | Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin 1 from Rattus norvegicus | |
| 2 | View Details | [314..508] | 1.231963 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [509..633] | 17.221849 | Dynamin | |
| 4 | View Details | [634..720] | 17.568636 | No description for PF02212.9 was found. Confident ab initio structure predictions are available. | |
| 5 | View Details | [721..877] | 31.69897 | No description for 1y0fB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)