













| General Information: |
|
| Name(s) found: |
Tango4-PA /
FBpp0073459
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 482 amino acids |
Gene Ontology: |
|
| Cellular Component: |
catalytic step 2 spliceosome
[IDA]
precatalytic spliceosome [IDA] cytosol [IDA] |
| Biological Process: |
Golgi organization
[IMP]
nuclear mRNA splicing, via spliceosome [IC] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMEDVQKHS VHTLIFRSLK RTHDLFVSNQ GNLPEIDERL EKLRRSIKAK DGYGLVLDKV 60
61 AKIGEARKAS GAQTPGDKTL AITDGSATDA ASGAGSLVKY NAGARPAEKT APLVTGTHTS 120
121 LVRASLANSN ADMSANGAIA SHLQLIPKKA PSIPKPKWHA PWKLSRVISG HLGWVRCIAV 180
181 EPGNEWFATG AGDRVIKIWD LASGKLKLSL TGHVSTVRGV AVSTKHPYLF SCGEDRQVKC 240
241 WDLEYNKVIR HYHGHLSAVY SLALHPTIDV LATSGRDSTA RIWDMRTKAN VHTLTGHTNT 300
301 VASVVAQATN PQIITGSHDS TVRLWDLAAG KSVCTLTNHK KSVRSIVLHP SLYMFASASP 360
361 DNIKQWRCPE GKFVQNISGH TSIVNCMAAN SEGVLVSGGD NGTMFFWDWR TGYNFQRFQA 420
421 PVQPGSMDSE AGIFAMCFDQ SGSRLITAEA DKTIKVYKED DEASEESHPI NWRPDLLKRR 480
481 KF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
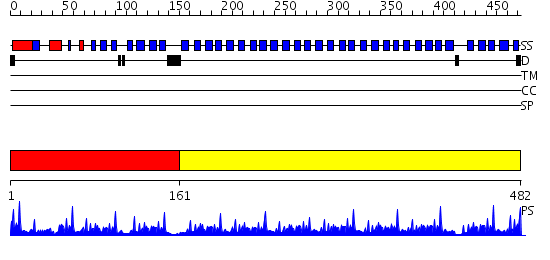
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | 8.221849 | Tricorn protease; Tricorn protease N-terminal domain; Tricorn protease domain 2 | |
| 2 | View Details | [161..482] | 80.69897 | PAF-AH Holoenzyme: Lis1/Alfa2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)