













| General Information: |
|
| Name(s) found: |
rdgA-PA /
FBpp0291800
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1457 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[NAS]
|
| Biological Process: |
actin filament organization
[IMP]
phosphoinositide biosynthetic process [TAS] visual perception [NAS] activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway [IEA] phototransduction [IMP][TAS] lipid phosphorylation [TAS] phosphatidic acid biosynthetic process [TAS] intracellular signaling pathway [IEA] thermotaxis [IDA] photoreceptor cell maintenance [NAS][TAS] rhodopsin mediated signaling pathway [IMP] phosphorylation [NAS] deactivation of rhodopsin mediated signaling [IGI] |
| Molecular Function: |
diacylglycerol binding
[ISS][NAS]
diacylglycerol kinase activity [ISS][NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQQQQQPSID QLPEPTASTS NSATTKPTIA TATTSTTTTS GNNFHQQLQA TTAATMQRLR 60
61 TTFTRSRTPT GAEMKMQNSL EVPKQVRSAS FDEMQLESQR ASSSLLKQQS SSSASADERS 120
121 SEAGFLQVPL AAHQQRSHSF DSATASAGSD DSGTFLEVPR RLKARRSSST KTPPPCIHCH 180
181 YLEEYERRMT AEQRYFIDHR ELTALSYSNT SSEASEDEDE VEGHNAEEEE EGSAAIEDAE 240
241 EETTEAATEE ADEDPRTEVE SEHDHDPDDD AALEMDIRIG NMSQGSSIEE SRARLPRQMR 300
301 RHTIGSSSVT SASEDEGLEG SDNGSPHFGN TLLPPQPTTP CGITFTLSPT NGDYPSPPHL 360
361 PLDPGSPPIS PCSSNSGRLP ALAPIISTPC SSADADDAGA AMGLPVRARR RSISRQEAIF 420
421 VEPTGNSLEN VSHEEVDNSN TKSSVDTADS LDEASTMATC GSPGAAGGSG ASSSHHNAFV 480
481 VRDIYLMVPD LKRDRAASVD SCFSKLSSNA KTEELQPSAD GCFLTVPNIN ATRSRSVDIV 540
541 LPTDEQARYK ALSMTGSTVT YADGRTASAS NSRRPIRIVP DWTENAVQGE HYWKPTSASG 600
601 DLCCLNEECI KSGQRMKCSA CQLVAHHNCI PFVNEKSTLA CKPTYRDVGI RQYREQTTTH 660
661 HHWVHRKLEK GKCKQCGKFF PMKQAVQSKL FGSKEIVALA CAWCHEIYHN KEACFNQAKI 720
721 GEECRLGNYA PIIVPPSWIV KLPTKGNFKS SIRVSNKNNA ASGSGGGGAG GGAGGGGGKS 780
781 KKQTQRRQKG KEEKKEPRAF IVKPIPSPEV IPVIVFINPK SGGNQGHKLL GKFQHLLNPR 840
841 QVFDLTQGGP KMGLDMFRKA PNLRVLACGG DGTVGWVLSV LDQIQPPLQP APAVGVLPLG 900
901 TGNDLARALG WGGGYTDEPI GKILREIGMS QCVLMDRWRV KVTPNDDVTD DHVDRSKPNV 960
961 PLNVINNYFS FGVDAHIALE FHEAREAHPE RFNSRLRNKM YYGQMGGKDL ILRQYRNLSQ1020
1021 WVTLECDGQD FTGKLRDAGC HAVLFLNIPS YGGGTHPWND SFGASKPSID DGLMEVVGLT1080
1081 TYQLPMLQAG MHGTCICQCR KARIITKRTI PMQVDGEACR VKPSVIEIEL LNKALMLSKR1140
1141 KHGRGDVQVN PLEKMQLHIL RVTMQQYEQY HYDKEMLRKL ANKLGQIEIE SQCDLEHVRN1200
1201 MLNTKFEESI SYPKVSQDWC FIDSCTAEHY FRIDRAQEHL HYICDIAIDE LYILDHEAAT1260
1261 MPQTPDQERS FAAFSQRQAQ NERRQMDQAQ GRGPGSTDED LQIGSKPIKV MKWKSPILEQ1320
1321 TSDAILLAAQ SGDLNMLRAL HEQGYSLQSV NKNGQTALHF ACKYNHRDIV KYIIASATRR1380
1381 LINMADKELG QTALHIAAEQ NRRDICVMLV AAGAHLDTLD SGGNTPMMVA FNKNANEIAT1440
1441 YLESKQGTQP VDGWLDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
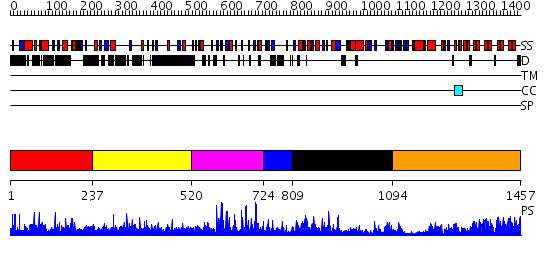
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..236] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [237..519] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [520..723] | 45.0 | No description for 2i13A was found. | |
| 4 | View Details | [724..808] | 15.522879 | Transcription factor IIIA, TFIIIA | |
| 5 | View Details | [809..1093] | 4.33 | No description for 2bonA was found. | |
| 6 | View Details | [1094..1457] | 84.522879 | Ankyrin-R |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)