













| General Information: |
|
| Name(s) found: |
FBpp0304835
[FlyBase]
arm-PD / FBpp0089035 [FlyBase] arm-PA / FBpp0089034 [FlyBase] arm-PB / FBpp0089032 [FlyBase] arm-PE / FBpp0089031 [FlyBase] |
| Description(s) found:
Found 73 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 843 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apicolateral plasma membrane
[TAS]
nucleus [IPI] adherens junction [IDA][NAS][TAS] spot adherens junction [NAS] cytoplasm [IDA][NAS][TAS] zonula adherens [IDA][NAS][TAS] membrane [NAS] catenin complex [ISS][NAS] apical plasma membrane [IDA] |
| Biological Process: |
dorsal closure
[IMP][TAS]
regulation of cell shape [IMP] ventral furrow formation [IMP] wing disc morphogenesis [IMP] protein localization [TAS] nervous system development [IMP] germ-line stem cell division [NAS] positive regulation of transcription [IMP] cell adhesion [IMP] cytoskeletal anchoring at plasma membrane [ISS] heart development [NAS][TAS] compound eye morphogenesis [IMP] ovarian follicle cell development [TAS] somatic stem cell maintenance [IMP] Wnt receptor signaling pathway [IMP][ISS][NAS][TAS] positive regulation of gene-specific transcription from RNA polymerase II promoter [IDA] neuroblast development [IMP] cuticle pattern formation [IMP] oocyte localization during germarium-derived egg chamber formation [TAS] cell-cell adhesion [IMP] delamination [IMP] cell morphogenesis [IMP] establishment or maintenance of cell polarity [NAS] imaginal disc-derived wing expansion [IMP] compound eye retinal cell programmed cell death [IMP] neuroblast fate determination [NAS] photoreceptor cell differentiation [IMP] oogenesis [IMP] positive regulation of JNK cascade [TAS] zonula adherens assembly [IMP][TAS] branch fusion, open tracheal system [IMP] |
| Molecular Function: |
alpha-catenin binding
[NAS]
protein binding [IPI] transcription coactivator activity [IDA] transcription activator activity [IDA] cytoskeletal protein binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYMPAQNRT MSHNNQYNPP DLPPMVSAKE QTLMWQQNSY LGDSGIHSGA VTQVPSLSGK 60
61 EDEEMEGDPL MFDLDTGFPQ NFTQDQVDDM NQQLSQTRSQ RVRAAMFPET LEEGIEIPST 120
121 QFDPQQPTAV QRLSEPSQML KHAVVNLINY QDDAELATRA IPELIKLLND EDQVVVSQAA 180
181 MMVHQLSKKE ASRHAIMNSP QMVAALVRAI SNSNDLESTK AAVGTLHNLS HHRQGLLAIF 240
241 KSGGIPALVK LLSSPVESVL FYAITTLHNL LLHQDGSKMA VRLAGGLQKM VTLLQRNNVK 300
301 FLAIVTDCLQ ILAYGNQESK LIILASGGPN ELVRIMRSYD YEKLLWTTSR VLKVLSVCSS 360
361 NKPAIVDAGG MQALAMHLGN MSPRLVQNCL WTLRNLSDAA TKVEGLEALL QSLVQVLGST 420
421 DVNVVTCAAG ILSNLTCNNQ RNKATVCQVG GVDALVRTII NAGDREEITE PAVCALRHLT 480
481 SRHVDSELAQ NAVRLNYGLS VIVKLLHPPS RWPLIKAVIG LIRNLALCPA NHAPLREHGA 540
541 IHHLVRLLMR AFQDTERQRS SIATTGSQQP SAYADGVRME EIVEGTVGAL HILARESHNR 600
601 ALIRQQSVIP IFVRLLFNEI ENIQRVAAGV LCELAADKEG AEIIEQEGAT GPLTDLLHSR 660
661 NEGVATYAAA VLFRMSEDKP QDYKKRLSIE LTNSLLREDN NIWANADLGM GPDLQDMLGP 720
721 EEAYEGLYGQ GPPSVHSSHG GRAFHQQGYD TLPIDSMQGL EISSPVGGGG AGGAPGNGGA 780
781 VGGASGGGGN IGAIPPSGAP TSPYSMDMDV GEIDAGALNF DLDAMPTPPN DNNNLAAWYD 840
841 TDC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
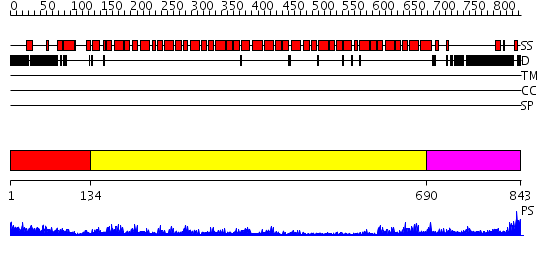
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 77.69897 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP | |
| 2 | View Details | [134..689] | 93.522879 | No description for 2gl7A was found. | |
| 3 | View Details | [690..843] | 20.0 | Importin beta |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)