













| General Information: |
|
| Name(s) found: |
Gnf1-PA /
FBpp0078478
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 986 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
DNA replication factor C complex [IEA][NAS] |
| Biological Process: |
DNA replication
[IEA][NAS]
|
| Molecular Function: |
DNA binding
[IDA][NAS]
ATP binding [IEA] DNA clamp loader activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQRGIDSFFK RLPAKAKSAE AENGETPSKA PKRRKAVIIS SDEDEVVSPP ETKKRKASKT 60
61 ASSEDDVVAA TPEPIAKKAR NGQKPALSKL KRHVDPTELF GGETKRVIVP KPKTKAVLEF 120
121 ENEDIDRSLM EVDLDESIKE AAPEKKVHSI TRSSPSPKRA KNSSPEPPKP KSTKSKATTP 180
181 RVKKEKPAAD LESSVLTDEE RHERKRASAV LYQKYKNRSS CLNPGSKEIP KGSPDCLSGL 240
241 TFVVTGVLES MEREEAESVI KEYGGKVMTV VGKKLKYLVV GEEAGPKKLA VAEELNIPIL 300
301 SEDGLFDLIR EKSGIAKQVK EEKKSPKKEH SSEEKGKKEV KTSRRSSDKK EKEATKLKYG 360
361 EKHDIAKHKV KEEHTSPKET KDKLNDVPAV TLKVKKEPSS QKEHPPSPRT ADLKTLDVVG 420
421 MAWVDKHKPT SIKEIVGQAG AASNVTKLMN WLSKWYVNHD GNKKPQRPNP WAKNDDGSFY 480
481 KAALLSGPPG IGKTTTATLV VKELGFDAVE FNASDTRSKR LLKDEVSTLL SNKSLSGYFT 540
541 GQGQAVSRKH VLIMDEVDGM AGNEDRGGMQ ELIALIKDSS IPIICMCNDR NHPKIRSLVN 600
601 YCYDLRFQRP RLEQIKGKIM SICFKEKVKI SPAKVEEIIA ATNNDIRQSI NHIALLSAKE 660
661 DASQKSGQQV ATKDLKLGPW EVVRKVFTAD EHKHMSFADK SDLFFHDYSL APLFVQQNYL 720
721 QVLPQGNKKD VLAKVAATAD ALSLGDLVEK RIRANSAWSL LPTQAFFSSV LPGEHMCGHF 780
781 TGQINFPGWL GKNSKSGKRA RLAQELHDHT RVCTSGSRLS VRLDYAPFLL DNIVRPLAKD 840
841 GQEGVPAALD VMKDYHLLRE DLDSLVELTS WPGKKSPLDA VDGRVKAALT RSYNKEVMAY 900
901 SYSAQAGIKK KKSEAAGADD DYLDEGPGEE DGAGGHLSSE EDEDKDNLEL DSLIKAKKRT 960
961 TTSKASGGSK KATSSTASKS KAKAKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
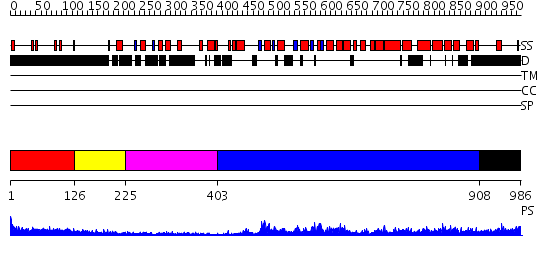
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..125] | 1.163993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [126..224] | 14.0 | Crystal structure of N-terminal domain of E.Coli Lon Protease | |
| 3 | View Details | [225..402] | 22.045757 | Crystal Structure Analysis of ClpB | |
| 4 | View Details | [403..907] | 54.69897 | Crystal Structure of the Eukaryotic Clamp Loader (Replication Factor C, RFC) Bound to the DNA Sliding Clamp (Proliferating Cell Nuclear Antigen, PCNA) | |
| 5 | View Details | [908..986] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)