













| General Information: |
|
| Name(s) found: |
DMAP1-PA /
FBpp0085546
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 433 amino acids |
Gene Ontology: |
|
| Cellular Component: |
NuA4 histone acetyltransferase complex
[IDA]
|
| Biological Process: |
phagocytosis, engulfment
[IMP]
negative regulation of transcription [IEA] |
| Molecular Function: |
DNA binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSADVRDILD MERANTPEVT RDSFLATKKR NFERTKTASR RPEGMHREVF ALLYTDKKDA 60
61 PPLLPTDTAL GIGAGYKETK ARLGMKKVRK WEWAPFSNPA RNDSAVFHHW KRVTDNSTDY 120
121 PFAKFNKQLE VPSYTMTEYN AHLRNNINNW SKVQTDHLFD LARRFDLRFI VMADRWNRQQ 180
181 HGTKTVEELK ERYYEVVALL AKAKNQTSEK KVFVYDVEHE RRRKEQLEKL FKRTTQQVEE 240
241 ENMLINEMKK IEARKKERER KTQDLQKLIS QADQQNEHAS NTPSTRKYEK KLHKKKVHQQ 300
301 PRPSRVDSVV NAIEIGSSGI KFADLRGSGV SLRSQRMKLP ANIGQRKVKA LEQAIQEFKV 360
361 DPAPPPTEDI CTSFNELRSD MVLLCELRTA LSTCVYEMES LKHQYEAACP GKTLNIPPSL 420
421 VPIKTEALDN STN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
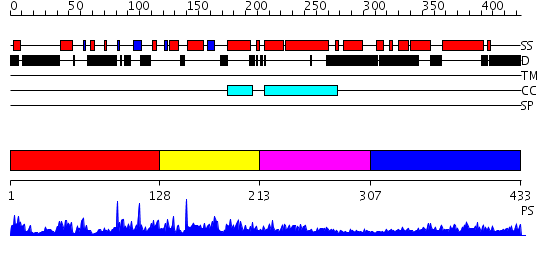
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | 3.040968 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [128..212] | 1.02 | Solution structures of the myb-like DNA binding domain of 4930532D21Rik protein | |
| 3 | View Details | [213..306] | 1.114985 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [307..433] | 3.108996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)