













| General Information: |
|
| Name(s) found: |
Strn-Mlck-PF /
FBpp0086412
[FlyBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1506 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein amino acid phosphorylation
[IEA][ISS][NAS]
|
| Molecular Function: |
ATP binding
[IEA]
calmodulin-dependent protein kinase activity [IDA] structural constituent of cytoskeleton [ISS] protein serine/threonine kinase activity [NAS] myosin light chain kinase activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHNIFFSIDI LNKSRMDKVH TRHGNIETLP RFVRNLRNLR CCDGDAISLE CHVEADPEPF 60
61 IIWEKDGHVM PSDRDYVMSF DGTKATLSIP RVYPEDEGEY TCVAKNSVGR SLSSACIIVD 120
121 VPEEKENMLS RQLARPSGLL SAHSTPRSTP RSTPARSFSP LRLSYRTSSI DLSGVAERRR 180
181 SDARNAITAP KFLAIPYNRV VEEGDSVRFQ CAISGHPTPW ATWDKDGLIV TPTPRIAVKE 240
241 IDDLRIIEID EVTFDDAGLY RVTLENDFGR IEATARLDVI RSSRYSKSPS VRSVRASSSR 300
301 RNAHLYRRIM GPSTAIGGRM ALASGYRGSS VPSVRFYHND VELEASERVH ILLQDSMALL 360
361 IVDNVTREDE GQYTCIISGD HDPLITSTTV TFHDSNTEIR RRRAVITERL PEITKSLEGE 420
421 VIDLCCSIEC DEPYSYVWLR NGEILPDSDE FNYIDHGNGR LCLRINDAFD IDSGIYSCQV 480
481 FTSDINDSTS DSTFDSHSIC SLINSGCSDC SSSGELCVLE RDLRGQDEEC VQLLKTPLPV 540
541 VCASGDEALF YARVFPCDAE ADWYLNGQLL AQADDSLNMT LESYPENGIR LLRMRDVTAS 600
601 RSGEICLQVK HPQAEFRRIP TTRTYTSLLV LPAIRGNSSS SSLAARSCIL TRPEDCTALI 660
661 GGHVRLSVRY EPFPGTKVIW YKACHPIVES SNVTIRTTSQ QSTLYITDIS ADDSGKYTVE 720
721 VMNDYGVEAA AASVAVEGPP EPPSGQPSVS LGPDRVAVAW CGPPYDGGCM ITGFIIEMQT 780
781 IGDENCDEDS WQQVTRVVDS LAYTVKNLQP ERQYRFRVRA ENIHGRSAPG QASELVQITN 840
841 TPQRSTSSDA SDRFGQATVS VQSGGDFKSR FEIIEELGKG RFGIVYKVQE RGQPEQLLAA 900
901 KVIKCIKSQD RQKVLEEISI MRALQHPKLL QLAASFESPR EIVMVMEYIT GGELFERVVA 960
961 DDFTLTEMDC ILFLRQVCDG VAYMHGQSVV HLDLKPENIM CHTRTSHQIK IIDFGLAQRL1020
1021 DTKAPVRVLF GTPEFIPPEI ISYEPIGFQS DMWSVGVICY VLLSGLSPFM GDTDVETFSN1080
1081 ITRADYDYDD EAFDCVSQEA KDFISQLLVH RKEDRLTAQQ CLASKWLSQR PDDSLSNNKI1140
1141 CTDKLKKFII RRKWQKTGNA IRALGRMANL SVSRRNSAIA MGVLSSPRPS ISGLGMLTTS1200
1201 AIGSGTSSQM TSLHEEEDDF SGEMPPVEKR TVLKLRDKSQ CSERSDSGYS ECSNCSGAQE1260
1261 TLLLSLAKSK LEAIAKASTL PTVVHDTEQP VSLELPTKGE AIMRSDFTNT IKMRKKSLED1320
1321 SAAREKPRSK PQVKPLLCES KLKVSQLKDR FQVSPAPASA SASASAANKP PLAYGPFKIA1380
1381 KVASVGRISR TEEPGRSGRG APSVSGKGKS PQVRSMPSSP LPQRSATPTR LMSQRVREAA1440
1441 ERLAQQHTVA SAQRHLGNGR GTGTGNGNGN SNSNGNGNGN TAETNRESRA RRLINRFNSE1500
1501 TQHITS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
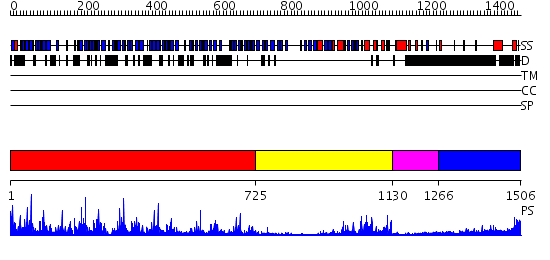
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..724] | 54.0 | Model of human carcinoembryonic antigen by homology modelling and curve-fitting to experimental solution scattering data | |
| 2 | View Details | [725..1129] | 34.39794 | CHICKEN SRC TYROSINE KINASE | |
| 3 | View Details | [1130..1265] | 46.045757 | TWITCHIN KINASE FRAGMENT (C.ELEGANS), AUTOREGULATED PROTEIN KINASE AND IMMUNOGLOBULIN DOMAINS | |
| 4 | View Details | [1266..1506] | 1.321997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)