













| General Information: |
|
| Name(s) found: |
FBpp0309018
[FlyBase]
dup-PA / FBpp0086519 [FlyBase] |
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 743 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[IDA]
nucleus [IDA] cytoplasm [IDA] nuclear origin of replication recognition complex [IDA][NAS] |
| Biological Process: |
DNA replication checkpoint
[IMP]
eggshell chorion gene amplification [TAS] DNA replication [IMP][TAS] DNA-dependent DNA replication [NAS] DNA endoreduplication [IMP] antimicrobial humoral response [IMP] chorion-containing eggshell formation [IMP] |
| Molecular Function: |
DNA binding
[NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQPSVAAFF TNRKRAALDD AISIKNRRLV EPAETVSPAS APSQLPAGDQ DADLDTLKAA 60
61 ATGMRTRSGR TARLIVTAAQ ESKKKTPAAA KMEPHIKQPK LVQFIKKGTL SPRKQAQSSK 120
121 LDEEELQQSS AISEHTPKVN FTITSQQNAD NVQRGLRTPT KQILKDASPI KADLRRQLTF 180
181 DEVKTKVSRS AKLQELKAVL ALKAALEQKR KEQEERNRKL RDAGPSPSKS KMSVQLKEFD 240
241 TIELEVLISP LKTFKTPTKI PPPTPDKHEL MSPRHTDVSK RLLFSPAKNG SPVKLVEVPA 300
301 YKRYASLVES SRAGQLPLPY KYRHLLDVFK GLDSVVAMFH NRKETITFKK LKPAVQRMLR 360
361 KNFTETHLAQ IKHIYPDAFI FSQVKTRNFG SVSKADYFQL IIAPNVEPLP EQQQSEKPQH 420
421 FTKINEDDVL ASAQSTSMNP HVMTARMQRF QSLLLDRAMR AHDQFLRSQD PPIIIEKALT 480
481 RWHPQFDLES CPEVELSPLP QPPNVEKYSS AKDILSTARN LFNCATPMER AMDRYEAKLE 540
541 SEKQQAAESN KKTEEQQAGE VTRTSTAIQT SQEVPGISGS SKNPTVPETT NQTETAKPTV 600
601 KDCTVPDASS NLLKGLPKSL IEKIRAKQAA KALEAMTRRP SQDQEATKYS RLPELARHLR 660
661 NVFVTERKGV LTLEVIIKKI QNSFRANLTP QEIEAHLKLL AKELPSWASF HEVRKTMYLK 720
721 VAKDMDMNKI IEKLESVANA KSN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
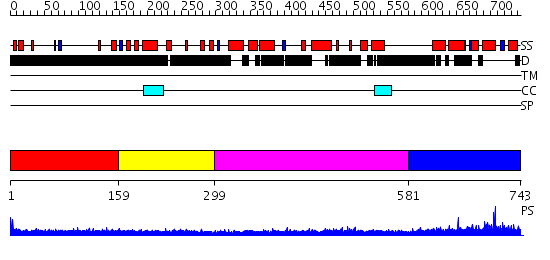
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..158] | 2.184995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [159..298] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [299..580] | 13.221849 | Strucure of Geminin-Cdt1 complex | |
| 4 | View Details | [581..743] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)