













| General Information: |
|
| Name(s) found: |
FBpp0294043
[FlyBase]
AGO1-PA / FBpp0086739 [FlyBase] AGO1-PC / FBpp0086738 [FlyBase] |
| Description(s) found:
Found 41 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 984 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
perinuclear region of cytoplasm [IDA] cytoplasm [IDA] RNA-induced silencing complex [ISS] eukaryotic translation initiation factor 2 complex [ISS] |
| Biological Process: |
synapse assembly
[TAS]
gene silencing by miRNA [IMP][NAS] production of miRNAs involved in gene silencing by miRNA [IMP] segment polarity determination [IGI] chromatin silencing [IMP] production of siRNA involved in RNA interference [IMP] mRNA cleavage involved in gene silencing by miRNA [IDA][IMP] mRNA catabolic process [IGI][IMP] RNA interference [IGI][TAS] cellularization [IGI] targeting of mRNA for destruction involved in RNA interference [IMP] |
| Molecular Function: |
miRNA binding
[IDA]
translation initiation factor activity [ISS] protein binding [IPI] enzyme binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTERELAPG GPAQLHPHTL PLTFPDLQMT STVGIIGKVY ESQWTPSPTR PQSPSQAQTS 60
61 FDTLTSPPAP GSSVNPTAVT SPSAQNVAAG GATVAGAAAT AAQVASALGA TTGSVTPAIA 120
121 TATPATQPDM PVFTCPRRPN LGREGRPIVL RANHFQVTMP RGYVHHYDIN IQPDKCPRKV 180
181 NREIIETMVH AYSKIFGVLK PVFDGRNNLY TRDPLPIGNE RLELEVTLPG EGKDRIFRVT 240
241 IKWQAQVSLF NLEEALEGRT RQIPYDAILA LDVVMRHLPS MTYTPVGRSF FSSPEGYYHP 300
301 LGGGREVWFG FHQSVRPSQW KMMLNIDVSA TAFYKAQPVI DFMCEVLDIR DINEQRKPLT 360
361 DSQRVKFTKE IKGLKIEITH CGQMRRKYRV CNVTRRPAQM QSFPLQLENG QTVECTVAKY 420
421 FLDKYRMKLR YPHLPCLQVG QEHKHTYLPL EVCNIVAGQR CIKKLTDMQT STMIKATARS 480
481 APDREREINN LVKRADFNND SYVQEFGLTI SNSMMEVRGR VLPPPKLQYG GRVSTGLTGQ 540
541 QLFPPQNKVS LASPNQGVWD MRGKQFFTGV EIRIWAIACF APQRTVREDA LRNFTQQLQK 600
601 ISNDAGMPII GQPCFCKYAT GPDQVEPMFR YLKITFPGLQ LVVVVLPGKT PVYAEVKRVG 660
661 DTVLGMATQC VQAKNVNKTS PQTLSNLCLK INVKLGGINS ILVPSIRPKV FNEPVIFLGA 720
721 DVTHPPAGDN KKPSIAAVVG SMDAHPSRYA ATVRVQQHRQ EIIQELSSMV RELLIMFYKS 780
781 TGGYKPHRII LYRDGVSEGQ FPHVLQHELT AIREACIKLE PEYRPGITFI VVQKRHHTRL 840
841 FCAEKKEQSG KSGNIPAGTT VDVGITHPTE FDFYLCSHQG IQGTSRPSHY HVLWDDNHFD 900
901 SDELQCLTYQ LCHTYVRCTR SVSIPAPAYY AHLVAFRARY HLVEKEHDSG EGSHQSGCSE 960
961 DRTPGAMARA ITVHADTKKV MYFA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
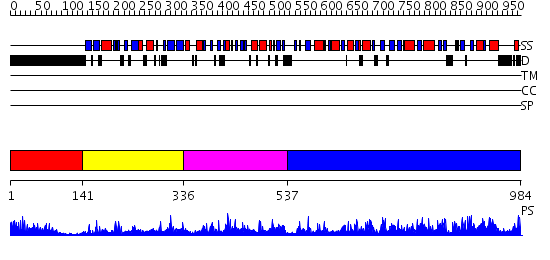
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | 7.69897 | No description for 1y0fA was found. | |
| 2 | View Details | [141..335] | 3.35 | Crystal structure of full length Argonaute from Pyrococcus furiosus | |
| 3 | View Details | [336..536] | 52.0 | Solution Structure of the Drosophila Argonaute 1 PAZ Domain | |
| 4 | View Details | [537..984] | 10.39794 | The Structure of a Piwi protein from Archaeoglobus fulgidus. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)