













| General Information: |
|
| Name(s) found: |
lin19-PD /
FBpp0087924
[FlyBase]
lin19-PA / FBpp0087923 [FlyBase] lin19-PB / FBpp0087922 [FlyBase] lin19-PC / FBpp0087921 [FlyBase] |
| Description(s) found:
Found 66 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 774 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear ubiquitin ligase complex
[ISS]
cullin-RING ubiquitin ligase complex [IEA] SCF ubiquitin ligase complex [NAS] |
| Biological Process: |
ubiquitin-dependent protein catabolic process
[IEA]
|
| Molecular Function: |
ubiquitin protein ligase binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNRSGNSQTT QKLVNLDDIW SELVEGIMQV FEHEKSLTRS QYMRFYTHVY DYCTSVSAAP 60
61 SGRSSGKTGG AQLVGKKLYD RLEQFLKSYL SELLTKFKAI SGEEVLLSRY TKQWKSYQFS 120
121 STVLDGICNY LNRNWVKREC EEGQKGIYKI YRLALVAWKG HLFQVLNEPV TKAVLKSIEE 180
181 ERQGKLINRS LVRDVIECYV ELSFNEEDTD AEQQKLSVYK QNFENKFIAD TSAFYEKESD 240
241 AFLSTNTVTE YLKHVENRLE EETQRVRGFN SKNGLSYLHE TTADVLKSTC EEVLIEKHLK 300
301 IFHTEFQNLL NADRNDDLKR MYSLVALSSK NLTDLKSILE NHILHQGTEA IAKCCTTDAA 360
361 NDPKTYVQTI LDVHKKYNAL VLTAFNNDNG FVAALDKACG KFINSNVVTI ANSASKSPEL 420
421 LAKYCDLLLK KSSKNPEDKE LEDNLNQVMV VFKYIEDKDV FQKYYSKMLA KRLVNHTSAS 480
481 DDAEAMMISK LKQTCGYEYT VKLQRMFQDI GVSKDLNSYF KQYLAEKNLT MEIDFGIEVL 540
541 SSGSWPFQLS NNFLLPSELE RSVRQFNEFY AARHSGRKLN WLYQMCKGEL IMNVNRNNSS 600
601 TYTLQASTFQ MSVLLQFNDQ LSFTVQQLQD NTQTQQENLI QVLQILLKAK VLTSSDNENS 660
661 LTPESTVELF LDYKNKKRRI NINQPLKTEL KVEQETVHKH IEEDRKLLIQ AAIVRIMKMR 720
721 KRLNHTNLIS EVLNQLSTRF KPKVPVIKKC IDILIEKEYL ERMEGHKDTY SYLA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
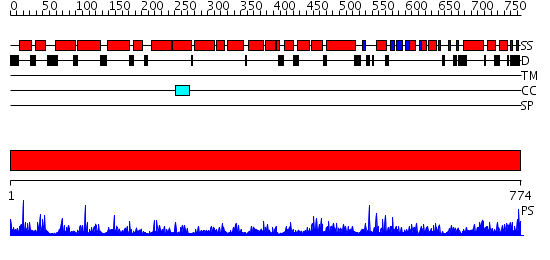
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..774] | 1000.0 | Crystal Structure of The Cand1-Cul1-Roc1 Complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)