













| General Information: |
|
| Name(s) found: |
mle-PA /
FBpp0085367
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1293 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
X chromosome [IDA] chromosome [NAS] chromatin [IDA] X chromosome located dosage compensation complex, transcription activating [NAS][TAS] |
| Biological Process: |
dosage compensation
[IMP][NAS][TAS]
axon extension [IMP] determination of adult lifespan [IMP] dosage compensation, by hyperactivation of X chromosome [NAS] male courtship behavior, veined wing generated song production [IGI] dosage compensation complex assembly involved in dosage compensation by hyperactivation of X chromosome [TAS] |
| Molecular Function: |
ATP binding
[IEA]
RNA helicase activity [NAS] double-stranded RNA binding [IEA][ISS] ATP-dependent helicase activity [IEA][TAS] helicase activity [ISS] chromatin binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDIKSFLYQF CAKSQIEPKF DIRQTGPKNR QRFLCEVRVE PNTYIGVGNS TNKKDAEKNA 60
61 CRDFVNYLVR VGKLNTNDVP ADAGASGGGP RTGLEGAGMA GGSGQQKRVF DGQSGPQDLG 120
121 EAYRPLNHDG GDGGNRYSVI DRIQEQRDMN EAEAFDVNAA IHGNWTIENA KERLNIYKQT 180
181 NNIRDDYKYT PVGPEHARSF LAELSIYVPA LNRTVTARES GSNKKSASKS CALSLVRQLF 240
241 HLNVIEPFSG TLKKKKDEQL KPYPVKLSPN LINKIDEVIK GLDLPVVNPR NIKIELDGPP 300
301 IPLIVNLSRI DSSQQDGEKR QESSVIPWAP PQANWNTWHA CNIDEGELAT TSIDDLSMDY 360
361 ERSLRDRRQN DNEYRQFLEF REKLPIAAMR SEILTAINDN PVVIIRGNTG CGKTTQIAQY 420
421 ILDDYICSGQ GGYANIYVTQ PRRISAISVA ERVARERCEQ LGDTVGYSVR FESVFPRPYG 480
481 AILFCTVGVL LRKLEAGLRG VSHIIVDEIH ERDVNSDFLL VILRDMVDTY PDLHVILMSA 540
541 TIDTTKFSKY FGICPVLEVP GRAFPVQQFF LEDIIQMTDF VPSAESRRKR KEVEDEEQLL 600
601 SEDKDEAEIN YNKVCEDKYS QKTRNAMAML SESDVSFELL EALLMHIKSK NIPGAILVFL 660
661 PGWNLIFALM KFLQNTNIFG DTSQYQILPC HSQIPRDEQR KVFEPVPEGV TKIILSTNIA 720
721 ETSITIDDIV FVIDICKARM KLFTSHNNLT SYATVWASKT NLEQRKGRAG RVRPGFCFTL 780
781 CSRARFQALE DNLTPEMFRT PLHEMALTIK LLRLGSIHHF LSKALEPPPV DAVIEAEVLL 840
841 REMRCLDAND ELTPLGRLLA RLPIEPRLGK MMVLGAVFGC ADLMAIMASY SSTFSEVFSL 900
901 DIGQRRLANH QKALSGTKCS DHVAMIVASQ MWRREKQRGE HMEARFCDWK GLQMSTMNVI 960
961 WDAKQQLLDL LQQAGFPEEC MISHEVDERI DGDDPVLDVS LALLCLGLYP NICVHKEKRK1020
1021 VLTTESKAAL LHKTSVNCSN LAVTFPYPFF VFGEKIRTRA VSCKQLSMVS PLQVILFGSR1080
1081 KIDLAANNIV RVDNWLNFDI EPELAAKIGA LKPALEDLIT VACDNPSDIL RLEEPYAQLV1140
1141 KVVKDLCVKS AGDFGLQRES GILPHQSRQF SDGGGPPKRG RFETGRFTNS SFGRRGNGRT1200
1201 FGGGYGNNGG GYGNNGGGYG NIGGGYGNNA GGYGNNGGYG NNGGGYRNNG GGYGNNGGGY1260
1261 GNKRGGFGDS FESNRGSGGG FRNGDQGGRW GNF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
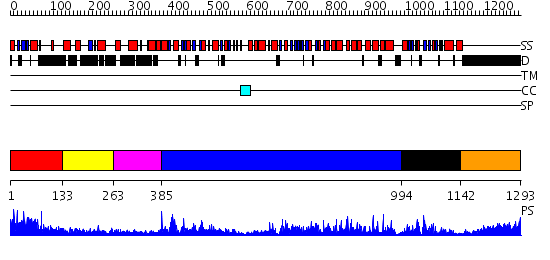
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | 48.0 | Solution structure of the N-terminal dsRBD from hypothetical protein BAB28848 | |
| 2 | View Details | [133..262] | 18.39794 | Double-stranded RNA-binding motif of Hypothetical protein BAB28848 | |
| 3 | View Details | [263..384] | 8.0 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [385..993] | 19.0 | No description for 2p6rA was found. | |
| 5 | View Details | [994..1141] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1142..1293] | 16.30103 | No description for 1y0fB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|