













| General Information: |
|
| Name(s) found: |
YJVE_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1279 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSYDFQKLT NDENLQGTES QVPSGSKSAS TNGLLSAASS AAGSSFGLTP SAILQQKHEN 60
61 AQQGKKQNNS KSFSKKPAID VHSEDAFPTL LSKTGPSKPR IVSWVRKTAS NTSVAGSDSV 120
121 SRDKIPFSAS SRASSTKSTL SSVKETDFVT ETLILSPDNQ APRMSFVGKP NSVAEIVRTV 180
181 MHQTSTRINV STASKTKNTT FLIQGKTSAV KAARRQILKL IGRRETKTMP CPVFVVGAII 240
241 GTNGQNLKSI MDRTSTRIQI PKRNNTANES SDDAKKPEKE ENSAASTLDD LEPQYEMTTI 300
301 TIEGDFEGVE LAQKDIEAII NERTSNTTVR ISHISTELYS LLRGPDGKNI KELEEGRDLK 360
361 VQIPFAYLDP SAPVNPIVLS GEKSAVRECA LYLQGQAEEL LRTTIPTMLP IPRRQHRFIN 420
421 GEKGVGIQDI LRKSGCSVIL PPINGDSDVV SVRGPALNIS EGIRLTLERA NSTIVDAVNI 480
481 TTAYASSKNP FDIASIVARL FLRSRKLIPL EEECAVQYHL PKREELQSNS NKTVIIEISG 540
541 KSQEAVREGR AKLLALVNQF PESKFYKVTI DPLLQRYVIG SKGKNLQKLR NEHQVELLVG 600
601 EYGEEDPDVI VCYIGADDGK SPDQIQKELA DLAESVKSSA EASAKIVSEI IQVPSVYHKH 660
661 IVGPKGTTLN AIIGKSEENV IVQLGKVSYR PDSTDDDVYI RGFSKDVERV VSEIKQVVRD 720
721 AKNHEILHSH VEEFDFPAQY SKNVIGKNGS NVSSLREDLG VQINVEEGHI RIQGIKKNVE 780
781 ETAARIKSQI EALIDDTILR VNIPNDFHRQ LIGSNGKYVR RLEEKFSVRV RFPREDDSSN 840
841 STGNELMKPT SPDEVVIRGG KKSVAAAKQE LLELYEYEKS IAYTSTIDIP SKAVSRVVGR 900
901 NGSTVENIRT QFDVKIDIGD VSTEETTPVS VRGAKADVEN AIKEISAIAE EVKNLVEKVI 960
961 KIDREYHRYL IGPNGSKLQN TIKECGGSTD KTETARLISF SNGNSEEERN SVVLRGDKEI1020
1021 VEALETRLLE IVEELKNQVE EKIEVPQRCI SSIIGRMGST RRDIERKTST MLNIPNVLDP1080
1081 EETVTITIVG SPENCEKAKE MIQEKVASQY TQMITVPDTV YESIMKGILM KKLRSDLKVF1140
1141 VDTPEIKPVQ PTEVVLEDHE DGVFPWKLVT HDYTGSSSSE WAVRGHKENV EKAIASLEKS1200
1201 IKQVMENCIA YLGIPTNLHR RIIGSGGSII NKIRKIAQVK IDVPRTPGDE IVVVQGSRAG1260
1261 VVKAKDLIFE RLQENQNQE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
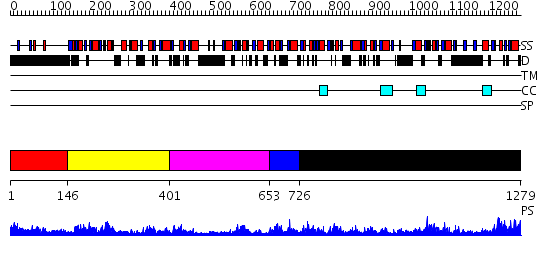
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [146..400] | 20.0 | No description for 2annA was found. | |
| 3 | View Details | [401..652] | 4.5 | No description for 2annA was found. | |
| 4 | View Details | [653..725] | 3.30103 | Structural genomics, 1.5A crystal structure of a hypothetical protein APE0754 from Aeropyrum pernix | |
| 5 | View Details | [726..1279] | 66.221849 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)