













| General Information: |
|
| Name(s) found: |
gi|109072241
[NCBI NR]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Macaca mulatta |
| Length: | 760 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGQQFQYDD SGNTFFYFLT SFVGLIVIPA TYYLWPRDQN AEQIRLKNIR KVYGRCMWYR 60
61 LRLLKPQPNI IPTVKKIVLL AGWALFLFLA YKVSKTDREY QEYNPYEVLN LDPGATVAEI 120
121 KKQYRLLSLK YHPDKGGDEV MFMRIAKAYA ALTDEESRKN WEEFGNPDGP QATSFGIALP 180
181 AWIVDQKNSI LVLLVYGLAF MVILPVVVGS WWYRSIRYSG DQILIRTTQI YTYFVYKTRN 240
241 MDMKRLIMVL AGASEFDPQY NKDATSRPTD NILIPQLIRE IGSINLKKNE PPLTCPYSLK 300
301 ARVLLLSHLA RMKIPETLEE DQQFMLKKCP ALLQEMVNVI CQLIVMARNR EEREFRAPTL 360
361 ASLENCMKLS QMAVQGLQQF KSPLLQLPHI EEDNLRRVSN HKKYKIKTIQ DLVSLKESDR 420
421 HTLLHFLEDE KYEEVMAVLG SFPYVTMDIK SQVLDDEDSN NITVGSLVTV LVKLTRQTMA 480
481 EVFEKEQSIC AAEEQPAEDG QGETNKNRTK GGWQQKSKGP KKTAKSKKKK PLKKKPTPVL 540
541 LPQSKQQKQK QANGVVGNEA AVKEDEEEVS DKGSDSEEEE TNRDSQSEKD DGSDRDSDRE 600
601 QDEKQNKDDE AEWQELQQSI QRKERALLET KSKITHPVYS LYFPEEKQEW WWLYIADRKE 660
661 QTLISMPYHV CTLKDTEEVE LKFPAPGKPG NYQYTVFLRS DSYMGLDQIK PLKLEVHEAK 720
721 PVPENHPQWD TAIEGDEDQE DSEGFEDSFE EEEEEEEDDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
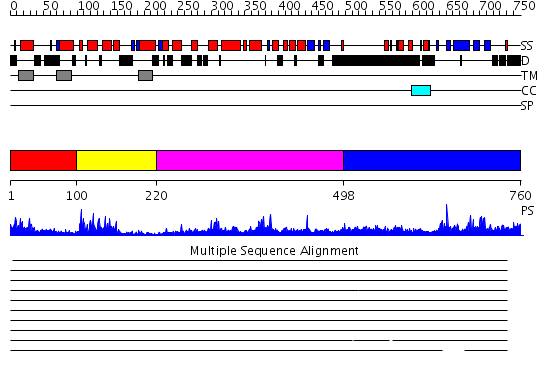
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..219] | 20.30103 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [220..497] | 56.69897 | No description for 2q0zX was found. | |
| 4 | View Details | [498..760] | 3.12 | No description for 2q0zX was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|