













| General Information: |
|
| Name(s) found: |
MCM3_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 879 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTELLADEVF KDRVRIFQEY LEHDTDDANV TLYQEAILRM LNMGQRRLIV NIDELRDYNR 60
61 ELADGVLLKP LEYVEPFDEA LRNVVSTLID PVVHKDLKDK LFYVGFRGSF GDHHVNPRTL 120
121 RAMHLNKMIS LEGIVTRCSF VRPKVIKSVH YCEATKRHHF KQYADATMNG GLSFQSTVYP 180
181 TQDENGNPLS IEFGFSTFRD HQSISLQEMP ERAPPGQLPR SIDILLDDDL VDTVKPGDRV 240
241 NIVGQYRSMG SKTSGNTSAT FRTVLLANNV VLLGNKPGLG NVGGGALDIT DADIRNINKL 300
301 ARKKNVFELL STSLAPSIYG YEYVKQAILL LLLGGTEKNL TNGTHIRGDI NILMVGDPST 360
361 AKSQLLRFVL NTAPLAIATT GRGSSGVGLT AAVTTDKETG ERRLEAGAMV LADRGVVCID 420
421 EFDKMSDIDR VAIHEVMEQQ TVTIAKAGIH TSLNARCSVI AAANPIYGQY DIRKDPHQNI 480
481 ALPDSMLSRF DLLFIVTDDI DDKKDRALSE HVLRMHRYLP PGVEPGTPVR DSLNSVLNVG 540
541 ATNAAGVSTE NVEQEVETPV WETFSSLLHA NARTKKKELL NINFVRKYIQ YAKSRIHPIL 600
601 NQATAEYITN IYCGLRNDDL QGNQRRTSPL TARTLETLIR LSTAHAKARL SSVVEVKDAK 660
661 AAEKILRYAL FREVVKPKRK KHKKQRLEAG EEFDSEDDNS DDMDIEESEE EMDTNMVIDS 720
721 GSRRVTRSQN ATSQSQESGS EIGSSIAGTA GSYNVGTSNT QLSWPSTHST LPATSRELAS 780
781 SDRNINTGTS VASEVSASVS EQSTVSLPRE KMSVFMARLA SLTKSELFSE ECASLEDVLE 840
841 SINNIEDDVG FSREEAIVAL KEMDAQNKIM FSDNVVYRI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
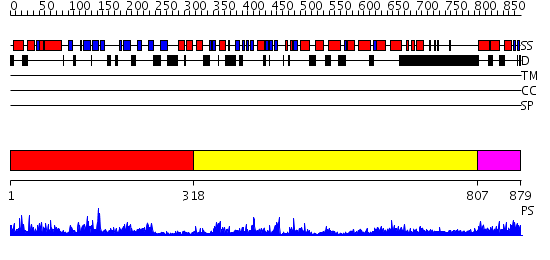
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | 40.0 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 2 | View Details | [318..806] | 50.522879 | EDTA treated | |
| 3 | View Details | [807..879] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)