













| General Information: |
|
| Name(s) found: |
MCM7
[HGNC (HUGO)]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Homo sapiens |
| Length: | 719 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALKDYALEK EKVKKFLQEF YQDDELGKKQ FKYGNQLVRL AHREQVALYV DLDDVAEDDP 60
61 ELVDSICENA RRYAKLFADA VQELLPQYKE REVVNKDVLD VYIEHRLMME QRSRDPGMVR 120
121 SPQNQYPAEL MRRFELYFQG PSSNKPRVIR EVRADSVGKL VTVRGIVTRV SEVKPKMVVA 180
181 TYTCDQCGAE TYQPIQSPTF MPLIMCPSQE CQTNRSGGRL YLQTRGSRFI KFQEMKMQEH 240
241 SDQVPVGNIP RSITVLVEGE NTRIAQPGDH VSVTGIFLPI LRTGFRQVVQ GLLSETYLEA 300
301 HRIVKMNKSE DDESGAGELT REELRQIAEE DFYEKLAASI APEIYGHEDV KKALLLLLVG 360
361 GVDQSPRGMK IRGNINICLM GDPGVAKSQL LSYIDRLAPR SQYTTGRGSS GVGLTAAVLR 420
421 DSVSGELTLE GGALVLADQG VCCIDEFDKM AEADRTAIHE VMEQQTISIA KAGILTTLNA 480
481 RCSILAAANP AYGRYNPRRS LEQNIQLPAA LLSRFDLLWL IQDRPDRDND LRLAQHITYV 540
541 HQHSRQPPSQ FEPLDMKLMR RYIAMCREKQ PMVPESLADY ITAAYVEMRR EAWASKDATY 600
601 TSARTLLAIL RLSTALARLR MVDVVEKEDV NEAIRLMEMS KDSLLGDKGQ TARTQRPADV 660
661 IFATVRELVS GGRSVRFSEA EQRCVSRGFT PAQFQAALDE YEELNVWQVN ASRTRITFV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
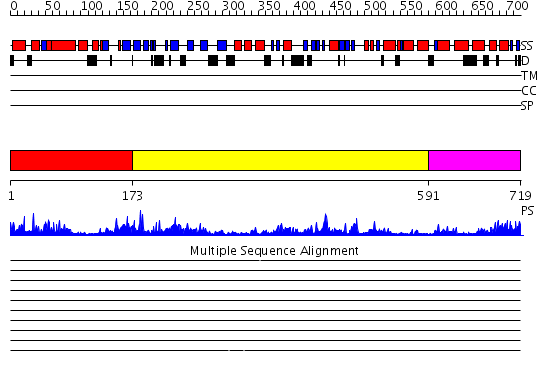
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | 35.39794 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 2 | View Details | [173..590] | 33.69897 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 3 | View Details | [591..719] | 47.69897 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)