













| General Information: |
|
| Name(s) found: |
RSC30 /
YHR056C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 883 amino acids |
Gene Ontology: |
|
| Cellular Component: |
RSC complex
[IDA |
| Biological Process: |
ATP-dependent chromatin remodeling
[IDA regulation of transcription, DNA-dependent [IMP double-strand break repair via nonhomologous end joining [IDA |
| Molecular Function: |
DNA binding
[ISS DNA-dependent ATPase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMDMQVRKVR KPPACTQCRK RKIGCDRAKP ICGNCVKYNK PDCFYPDGPG KMVAVPSASG 60
61 MSTHGNGQGS NHFSQGNGVN QKNVMIQTQY PIMQTSIEAF NFSFNPSVDT AMQWTKAASY 120
121 QNNNTNNNTA PRQNSSTVSS NVHGNTIVRS DSPDVPSMDQ IREYNTRLQL VNAQSFDYTD 180
181 NPYSFNVGIN QDSAVFDLMT SPFTQEEVLI KEIDFLKNKL LDLQSLQLKS LKEKSNLNAD 240
241 NTTANKINKT GENSKKGKVD GKRAGFDHQT SRTSQSSQKY FTALTITDVQ SLVQVKPLKD 300
301 TPNYLFTKNF IIFRDHYLFK FYNILHDICH INQFKVSPPN NKNHQQYMEV CKVNFPPKAI 360
361 IIETLNSESL NNLNIEEFLP IFDKTLLLEF VHNSFPNGDT CPSFSTVDLP LSQLTKLGEL 420
421 TVLLLLLNDS MTLFNKQAIN NHVSALMNNL RLIRSQITLI NLEYYDQETI KFIAITKFYE 480
481 SLYMHDDHKS SLDEDLSCLL SFQIKDFKLF HFLKKMYYSR HSLLGQSSFM VPAAENLSPI 540
541 PASIDTNDIP LIANDLKLLE TQAKLINILQ GVPFYLPVNL TKIESLLETL TMGVSNTVDL 600
601 YFHDNEVRKE WKDTLNFINT IVYTNFFLFV QNESSLSMAV QHSSNNNKTS NSERCAKDLM 660
661 KIISNMHIFY SITFNFIFPI KSIKSFSSGN NRFHSNGKEF LFANHFIEIL QNFIAITFAI 720
721 FQRCEVILYD EFYKNLSNEE INVQLLLIHD KILEILKKIE IIVSFLRDEM NSNGSFKSIK 780
781 GFNKVLNLIK YMLRFSKKKQ NFARNSDNNN VTDYSQSAKN KNVLLKFPVS ELNRIYLKFK 840
841 EISDFLMERE VVQRSIIIDK DLESDNLGIT TANFNDFYDA FYN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
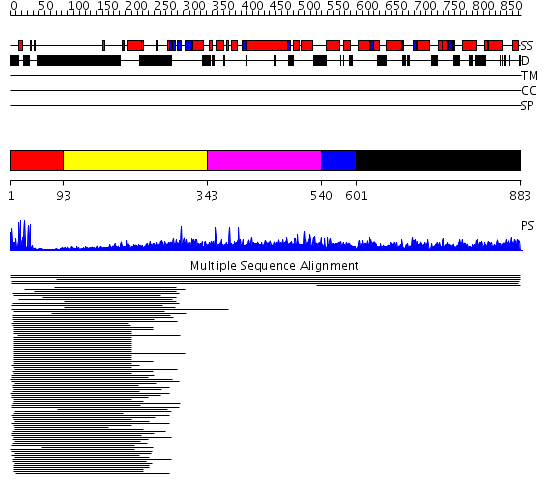
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | 6.09691 | PPR1 | |
| 2 | View Details | [93..342] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [343..539] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [540..600] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [601..883] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)