













| General Information: |
|
| Name(s) found: |
CSE4 /
YKL049C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 229 amino acids |
Gene Ontology: |
|
| Cellular Component: |
condensed nuclear chromosome, centromeric region
[IGI extrachromosomal circular DNA [IDA nuclear nucleosome [IDA kinetochore [IGI |
| Biological Process: |
mitotic sister chromatid segregation
[IMP 2-micrometer plasmid partitioning [IMP |
| Molecular Function: |
sequence-specific DNA binding
[IDA centromeric DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSKQQWVSS AIQSDSSGRS LSNVNRLAGD QQSINDRALS LLQRTRATKN LFPRREERRR 60
61 YESSKSDLDI ETDYEDQAGN LEIETENEEE AEMETEVPAP VRTHSYALDR YVRQKRREKQ 120
121 RKQSLKRVEK KYTPSELALY EIRKYQRSTD LLISKIPFAR LVKEVTDEFT TKDQDLRWQS 180
181 MAIMALQEAS EAYLVGLLEH TNLLALHAKR ITIMKKDMQL ARRIRGQFI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CBF1 CSE4 MIF2 |
| View Details | Qiu et al. (2008) |
|
CSE4 HSK3 MIF2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | No Comments | Wong J, et all (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
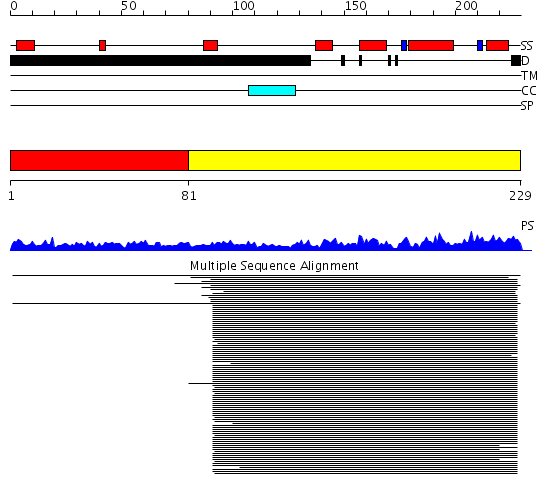
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..229] | 51.522879 | Histone H3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)