













| General Information: |
|
| Name(s) found: |
GCR2 /
YNL199C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 534 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
positive regulation of transcription from RNA polymerase II promoter
[IMP positive regulation of glycolysis [IMP |
| Molecular Function: |
transcription activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHHQTKLDVF IIRAYNLLSN ESVISGASLQ SVTNSPQTTT NTPSGMVNGA VGTGIANPTG 60
61 LMGSDSTPNI DEIITSTGSN ALTKTNSDSA NGTPNGNSSS TSAISNASNP ATTGNNASSS 120
121 ATSNGIYTQA QYSQLFAKIS KLYNATLSSG SIDDRSTSPK SAIELYQRFQ QMIKELELSF 180
181 DASPYAKYFR RLDGRLWQIK TDSELENDEL WRLVSMSIFT VFDPQTGQIL TQGRRKGNSL 240
241 NTSTKGSPSD LQGINNGNNN GNNGNIGNGS NIKNYGNKNM PNNRTKKRGT RVAKNAKNGK 300
301 NNKNSNKERN GITDTSAFSN TTISNPGTNM LFDPSLSQQL QKRLQTLSQD VNSRSLTGYY 360
361 TQPTSPGSGG FEFGLSHADL NPNASSNTMG YNTMSNNGSH SWKRRSLGSL DVNTLDDEAV 420
421 EELLQLTNTS KRQRPMTTAA EGALINDGPD TNLNANNTQM KVDLNPSNSM GPIDTEAVIR 480
481 PLKEAYDAII SEKGQRIVQL ERELELQRQE TQWLRKMLIE DMGCVRSMLR DLQR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
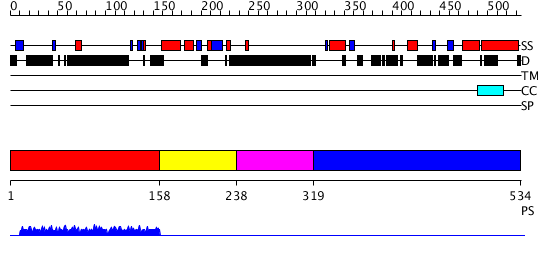
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 1.002946 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [158..237] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [238..318] | 1.002996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [319..534] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)