













| General Information: |
|
| Name(s) found: |
NMA111 /
YNL123W
[SGD]
|
| Description(s) found:
Found 37 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 997 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
lipid metabolic process
[IMP apoptosis [IMP |
| Molecular Function: |
serine-type peptidase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTISLSNIKK RDHSKISDGT SGESSLVKRK QLESATGDQE EEYTDHEIII EPLHFANNNN 60
61 TVLTDSENYL RWQNTISNVV KSVVSIHFSQ VAPFDCDSAL VSEATGFVVD AKLGIILTNR 120
121 HVVGPGPFVG YVVFDNHEEC DVIPIYRDPV HDFGFLKFDP KNIKYSKIKA LTLKPSLAKV 180
181 GSEIRVVGND AGEKLSILAG FISRIDRNAP EYGELTYNDF NTEYIQAAAS ASGGSSGSPV 240
241 VNIDGYAVAL QAGGSTEAST DFFLPLDRIL RALICIQTNK PITRGTIQVQ WLLKPYDECR 300
301 RLGLTSERES EARAKFPENI GLLVAETVLR EGPGYDKIKE GDTLISINGE TISSFMQVDK 360
361 IQDENVGKEI QLVIQRGGVE CTVTCTVGDL HAITPHRYVE VCGATFHELS YQMARFYALP 420
421 VRGVFLSSAS GSFNFDSKER VGWIVDSIDN KETPDLDTFI EIMKTIPDRK RVTVRYHHLT 480
481 DQHSPLVTSI YIDRHWCNEF RVYTRNDTTG IWDYKNVADP LPADALKPRS AKIIPIPVNN 540
541 EKVAKLSSSL CTVATMAAVP LDSLSADILK TSGLIIDAEK GYVLVSRRVV PHDCLDTFVT 600
601 IADSLVVPAT VEFLHPTHNF AIVKYDPELV KAPLITPKLS TTRMKRGDKL QFIGFTQNDR 660
661 IVTSETTVTD ISSVSIPSNL IPRYRATNLE AISIDCNVST RCNSGILTDN DGTVRGLWLP 720
721 FLGERLENKE KVYLMGLDIM DCREVIDILK NGGKPRVSIV DAGFGSISVL QARIRGVPEE 780
781 WIMRMEHESN NRLQFITVSR VSYTEDKIHL ETGDVILSVN GKLVTEMNDL NGVVSSADGI 840
841 LPSAMLDFKV VRDGNIVDLK IKTVEVQETD RFVIFAGSIL QKPHHAVLQA MVDVPKGVYC 900
901 TFRGESSPAL QYGISATNFI THVNEIETPD LDTFLKVVKT IPDNSYCKMR LMTFDNVPFA 960
961 ISLKTNYHYF PTAELKRDNI THKWIEKEFT GNSQSEK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
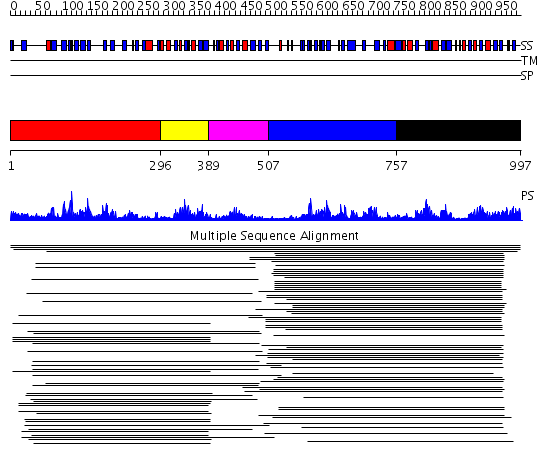
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..295] | 135.522879 | Mitochondrial serine protease HtrA2; Mitochondrial serine protease HtrA2, catalytic domain | |
| 2 | View Details | [296..388] | 135.522879 | Mitochondrial serine protease HtrA2; Mitochondrial serine protease HtrA2, catalytic domain | |
| 3 | View Details | [389..506] | 6.69897 | Photosystem II D1 C-terminal processing protease | |
| 4 | View Details | [507..756] | 156.201249 | Protease Do (DegP, HtrA), C-terminal domains; Protease Do (DegP, HtrA), catalytic domain | |
| 5 | View Details | [757..997] | 156.201249 | Protease Do (DegP, HtrA), C-terminal domains; Protease Do (DegP, HtrA), catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)