













| General Information: |
|
| Name(s) found: |
RRB1 /
YMR131C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 511 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
ribosome biogenesis
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKRSIEVNE EQDRVVSAKT ESHSVPAIPA SEEQDAPKND LEEQLSDEFD SDGEIIEIDG 60
61 DDEINDEDDL RKKQEEAETL VQKDQSEGNK EKIQELYLPH MSRPLGPDEV LEADPTVYEM 120
121 LHNVNMPWPC LTLDVIPDTL GSERRNYPQS ILLTTATQSS RKKENELMVL ALSNLAKTLL 180
181 KDDNEGEDDE EDDEDDVDPV IENENIPLRD TTNRLKVSPF AISNQEVLTA TMSENGDVYI 240
241 YNLAPQSKAF STPGYQIPKS AKRPIHTVKN HGNVEGYGLD WSPLIKTGAL LSGDCSGQIY 300
301 FTQRHTSRWV TDKQPFTVSN NKSIEDIQWS RTESTVFATA GCDGYIRIWD TRSKKHKPAI 360
361 SVKASNTDVN VISWSDKIGY LLASGDDNGT WGVWDLRQFT PSNADAVQPV AQYDFHKGAI 420
421 TSIAFNPLDE SIVAVGSEDN TVTLWDLSVE ADDEEIKQQA AETKELQEIP PQLLFVHWQK 480
481 EVKDVKWHKQ IPGCLVSTGT DGLNVWKTIS V |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
AMD1 RRB1 YJL070C |
| View Details | Qiu et al. (2008) |
|
DRS1 NAN1 NSR1 NUG1 PXR1 RRB1 UTP15 UTP4 UTP5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #33 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #35 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
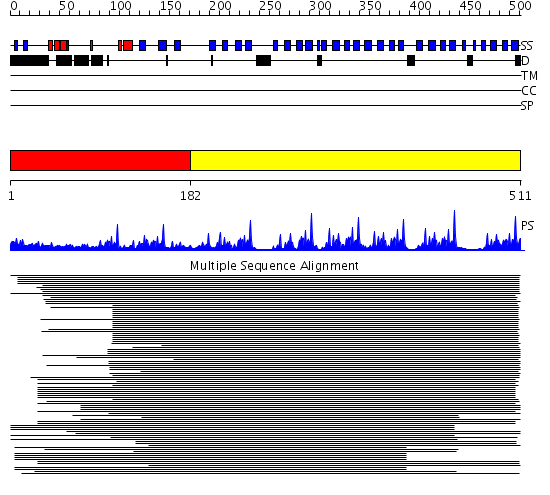
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | 1.673995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [182..511] | 87.70927 | beta1-subunit of the signal-transducing G protein heterotrimer |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)