













| General Information: |
|
| Name(s) found: |
BDS1 /
YOL164W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 646 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
dodecyl sulfate metabolic process
[IMP |
| Molecular Function: |
arylsulfatase activity
[IDA alkyl sulfatase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIGAFKRNRG SSQSFAKECQ PSTLKANLEV AKELPFSDRR DFEDATQGYI GSLSDEQIIG 60
61 PDGGVVWCMK SYGFLEPETP ANTVNPSLWR QAQLNAIHGL FKITDNVYQV RGLDISNMTI 120
121 IEGNTSLIII DTLFTTETAQ ESLKLYYRHR PQKPVRTVIY THSHSDHYGG VKGIVKEADV 180
181 KSGEVQIIAP VGFMESVVAE NILAGNAMHR RSQYQFGMLL SPSVKGHVDC GIGKAASHGT 240
241 VTLIAPTIII EEPVEERTID GVDFVFQLAP GSEAPSEMLI YMPQQRVLNM AEDVTHHMHN 300
301 LYALRGVEVR DGNQWAKYID AARVAFGSKT DVLIAQHHWP TTGQMRINEL LKKQRDMYKF 360
361 IHDQTLRLLN QGYTSRDIAE TLRMPSSLEQ EWSTRGYYGT LSHNVKAVYQ KYLGWYDANP 420
421 ANLNPLPPVA YAKKAVEYMG GADAVLARAY KDFQKGEFRW VASVVNQLVF ADPNNHQARE 480
481 LCADALEQLG YQAEASTWRN AYLVGAMELR QGVPKRRSTG KRNNIAVLNN EMFFDFLAVR 540
541 LNATKAEGKI IVSNWCFINS NERFVITLEN CALTYIQGWQ TDADATITLK RTTFEALLAN 600
601 EITMVDFLRS KEVEIEGNRL RIEELLKLFD DFDQSFPVVE PMGGST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
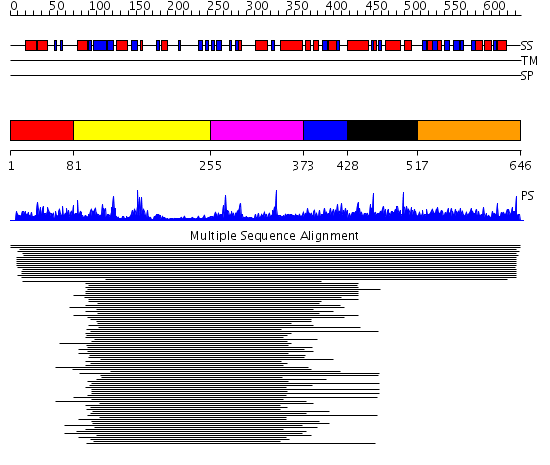
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..254] | 43.980454 | Zn metallo-beta-lactamase | |
| 3 | View Details | [255..372] | 43.980454 | Zn metallo-beta-lactamase | |
| 4 | View Details | [373..427] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [428..516] | 7.3 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 6 | View Details | [517..646] | 3.69897 | SCP2-like domain of MFE-2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 | No functions predicted. | ||||||||||||
| 5 | No functions predicted. | ||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)