













| General Information: |
|
| Name(s) found: |
ERG5 /
YMR015C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 538 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[TAS]
|
| Biological Process: |
ergosterol biosynthetic process
[TAS]
|
| Molecular Function: |
C-22 sterol desaturase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSVAENIIQ HATHNSTLHQ LAKDQPSVGV TTAFSILDTL KSMSYLKIFA TLICILLVWD 60
61 QVAYQIKKGS IAGPKFKFWP IIGPFLESLD PKFEEYKAKW ASGPLSCVSI FHKFVVIAST 120
121 RDLARKILQS SKFVKPCVVD VAVKILRPCN WVFLDGKAHT DYRKSLNGLF TKQALAQYLP 180
181 SLEQIMDKYM DKFVRLSKEN NYEPQVFFHE MREILCALSL NSFCGNYITE DQVRKIADDY 240
241 YLVTAALELV NFPIIIPYTK TWYGKKTADM AMKIFENCAQ MAKDHIAAGG KPVCVMDAWC 300
301 KLMHDAKNSN DDDSRIYHRE FTNKEISEAV FTFLFASQDA SSSLACWLFQ IVADRPDVLA 360
361 KIREEQLAVR NNDMSTELNL DLIEKMKYTN MVIKETLRYR PPVLMVPYVV KKNFPVSPNY 420
421 TAPKGAMLIP TLYPALHDPE VYENPDEFIP ERWVEGSKAS EAKKNWLVFG CGPHVCLGQT 480
481 YVMITFAALL GKFALYTDFH HTVTPLSEKI KVFATIFPKD DLLLTFKKRD PITGEVFE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
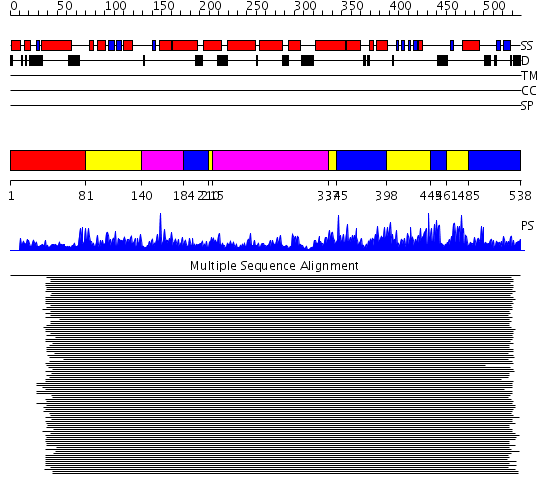
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..139] [210..214] [337..344] [398..444] [461..484] |
167.9691 | No description for 1jo9A_ was found. | |
| 3 | View Details | [140..183] [215..336] |
167.9691 | No description for 1jo9A_ was found. | |
| 4 | View Details | [184..209] [345..397] [445..460] [485..538] |
167.9691 | No description for 1jo9A_ was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 | |
| 2 | No functions predicted. |
| 3 | No functions predicted. |
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)