













| General Information: |
|
| Name(s) found: |
YLL032C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 825 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA ribosome [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNFKIYSTV ITTAFLQVPH LYTTNRLWKP IEAPFLVEFL QKRISSKELK NTKAICHIDP 60
61 SWVNLNASFI RDDMISIKAT TDDMDLDAIC RISLPLPMNT NDLTAELEKM KRILLDLSEK 120
121 FNLELIITKE PAYFTPEQTG ESKELCIYVH ALGFRSNLME CEPQLLAFVD LIKKNGMTLP 180
181 PQHYIIEPME LNSYSVLPLY MGVDMENFKH ISRAFKTSIY APSLITLSRD LKANPQIFFS 240
241 GAVHSLSLLA RKTLRESISV NSKSFFYRRL TNITPGKLLF IRKYYQQKVN QLILKYQSLI 300
301 RVTNEYIEFQ SISTNLLEMV IKNFTIQVLH EIVEVQISLN ENCAMSPELI IDSFFGHTGN 360
361 QIVVITPKED SFNQLIVVGN QSSTDEASDT SILHYLSDFI MGSNQVINPN LRQIKAIFEI 420
421 HPDFEDFISG KKNGKLTRIM ELSACLIQLE MEEEDDNLYL NLVSDSFPDF KESFKNVINE 480
481 FPAEESFFIP EVCHRPIIGT GGSLIQATMR KHNVFIQFSN SFNLPQNKIS MIRYDNVIIR 540
541 CPRKNKANIC LAKNDLKQIV QEYDSLQSKT LIRFSSGQYR HILHVNGQKN IIGQIEKNEN 600
601 VYIMIPLKEP LDGTSQLSIQ GNDENASRAA NELVNSAFGY EYEFKIDQEI DPNKEYEFYN 660
661 LIVVPFLQIM NIIVTFEKDL ITFTFEKDTN ENTLTKAIEL LSNYLETQKT KIIFKKIIKK 720
721 FVLGSASSKS NTSNSNTNGN FRSMNNAKSR TTIDNTSQSG ASPQRHKMPV ITTVGGAQAI 780
781 KGYIPNTYYN GYGYGYGYTY EYDYNYANSN KAQTNNRHKY QNGRK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
IRC8 LSM12 RPL38 YLL032C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
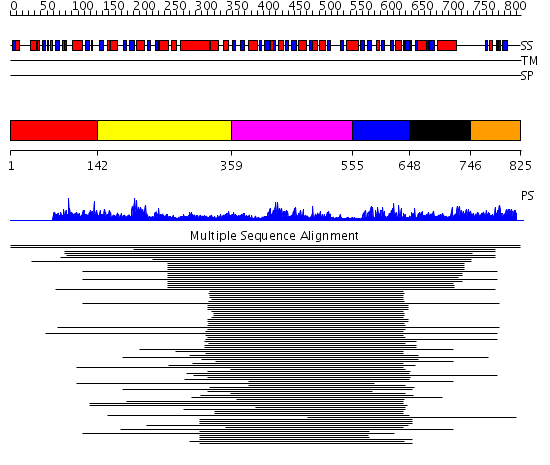
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [142..358] | 2.159989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [359..554] | 14.69897 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 4 | View Details | [555..647] | 19.69897 | Far upstream binding element, FBP, KH3 and KH4 domains | |
| 5 | View Details | [648..745] | 2.065989 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [746..825] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)