













| General Information: |
|
| Name(s) found: |
IRC8 /
YJL051W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 822 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCHNSVRSGN KAGFLGIKFG SALLSIATGA IAIALLCKFH DHEAVLIVIV CSTLLYGIPS 60
61 LISFITETVF APSKFHIGYF YNVLNFALPL ITMGCTVDYF HNTLRSPISV QSESHRVYIT 120
121 TLDSLLIFTL FINGIQLGFF LKDGNANNFG SSSNNISTDQ YDKEANAVEN GRFVPLKNSS 180
181 QTLTPDLELL HGSPKSMNGV AWLINELSTN SNTNANKTIS SDENSNSSVI RHKLGPISTS 240
241 KCPKKPSHSH FSKLKKYNSF FLGPKENRYK RNTQQATKVP TEKKSNHRSS QYVSRLSTIS 300
301 DISKSFLNFL ALNEKNGNST STARTPSEGR VSIIINEGNN TLKYKTPHDS HTIDSPNLEL 360
361 EREAIGRINS ALLPACLRVT DKMISPQQST QNEDSYQATP LIPQVEVDDD FYVGDILMTN 420
421 ELQDIPQVPR ISSDIEDDFE QQYTKHVDLP ARVTLEMWEK DQEKILQKVT TNRDKSKLLP 480
481 PFRFTSESDM DPSTSTELEV ELHAQNNFSF PFKSAGLQIA TSDQFNQQEF KTSDTISELD 540
541 EYLHDPSIQE EDASQLIESS LNQNNLSSTT IDNGPKDMSR FSTRHSPTKS IISMISGSGS 600
601 VKHQHSHSTL SNFFTGHSRN NSQINQLLQG SSSNMMSNTS PHSSPTKSLR MRFGKKLSLS 660
661 NISDTMSPYE GSTTDPINYS FGHGHNKNQS IDFSYVRTLQ SSHSPTKSTS GNSRRDSLNN 720
721 DRTQSTVNER ALRTASTLFY LQHNNATCTL NGEEPVLDTP QSIQSSSSGS EQESAGSGSG 780
781 YPEVVFSEYD REKWNVLRNL KEIAPEKTIE SGPVEELVSP SK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
IRC8 LSM12 RPL38 YLL032C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
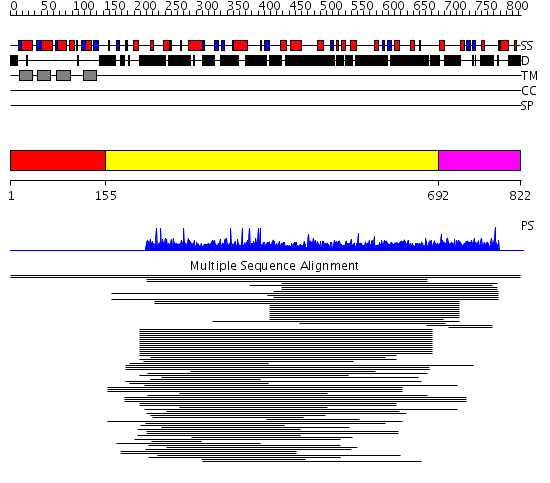
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..154] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [155..691] | 6.038997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [692..822] | 1.014984 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [480..548] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [549..678] | 1.11399 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [679..822] | 1.047983 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|