













| General Information: |
|
| Name(s) found: |
TTI1 /
YKL033W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1038 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSDTNAFKD IRISCVELSR IAFLPTESFD PNSLTLLACL KKVEEKLSAY EDDSLSPKFA 60
61 DYVFVPIASL LKQPALGESQ TEYVLLIIFH LLRTCWSSNG KFSEQLGQQL FPLITFLVSS 120
121 DKDNQKLITR SDEFKYAGCL VLHQFFKSVR SQRYHKEFFS NSKPNLLPAL GHSVTILLKI 180
181 LEQSPQNNEL QFKALASLEV LFQDIISDGE MLSFILPGNV SVFAKILTKP GRQIHYKVCV 240
241 RTLEVLAKLL VLVYDDFSLD IKVNKLTDIR ELSDTKLKHE INQSFMFNGP IVLLRTDGKT 300
301 HRDTSWLTAT SGQINIALEA FIPKLLKRNN ESIDEALATF VSILLTRCEN SLNNCEKVLV 360
361 STLVHLERDP MSKLPSHLVK LKEVVNEDLH KLSDIIRFEN ADRLSSLSFA ITILEKNNER 420
421 DTMINEVVRC LFESLNESIE PPSLINHKER IIEQSSQLTT TVNFENLEST NALIALPRLS 480
481 EDMSLKLKKF TYHMGSLLLE RHILNDVVTE LISEQVDSPR TQKIVALWLS TNFIKAMEKQ 540
541 PKEEEVYLQF ESDANYSSSM VEEVCLIVLE FCNELSQDIS MEIEGKGIKK SDEFAVCTVL 600
601 FSIETICAVM REEFQPELID YIYTVVDALA SPSEAIRYVS QSCALRIADT LYHGSIPNMI 660
661 LSNVDYLVES ISSRLNSGMT ERVSQILMVI CQLAGYETIE NFKDVIETIF KLLDYYHGYS 720
721 DLCLQFFQLF KIIILEMKKK YINDDEMILK IANQHISQST FSPWGMTDFQ QVLNILDKET 780
781 QVKDDITDEN DVDFLKDDNE PSNFQEYFDS KLREPDSDDD EEEREEEVEG SSKEYTDQWT 840
841 SPIPSDSYKI LLQILGYGER LLTHPSKRLR VQILIVMRLI FPLLSTQHNL LIREVASTWD 900
901 SIIQCVLCSD YSIVQPACSC VEQMIKYSGD FVAKRFIELW QKLCQDSFIL KELRIDPTVH 960
961 NHEKKSISKH VKFPPVTENA LVSMVHMVLE GVKITEYLIS EAVLEQIIYC CIQVVPVEKI1020
1021 SSMSLIVGDI VWKIRNIN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
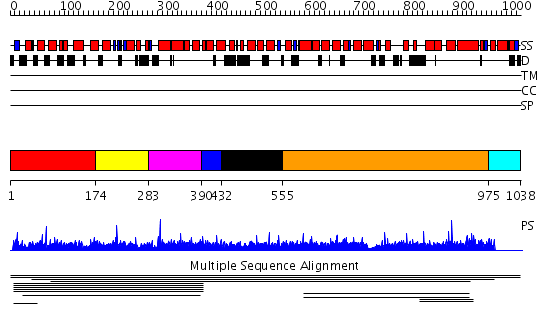
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] | 1.84 | beta-Catenin | |
| 2 | View Details | [174..282] | 1.84 | beta-Catenin | |
| 3 | View Details | [283..389] | 1.84 | beta-Catenin | |
| 4 | View Details | [390..431] | 1.84 | beta-Catenin | |
| 5 | View Details | [432..554] | 1.84 | beta-Catenin | |
| 6 | View Details | [555..974] | 2.006913 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [975..1038] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)