













| General Information: |
|
| Name(s) found: |
RSF2 /
YJR127C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1380 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
regulation of transcription
[IMP |
| Molecular Function: |
transcription factor activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPFAFGRGA PALCILTAAA RINLDNFVPC CWALFRLSFF FPLDPAYIRN ENKETRTSWI 60
61 SIEFFFFVKH CLSQHTFFSK TLAPKRNFRA KKLKDIGDTR IDRADKDFLL VPEPSMFVNG 120
121 NQSNFAKPAG QGILPIPKKS RIIKTDKPRP FLCPTCTRGF VRQEHLKRHQ HSHTREKPYL 180
181 CIFCGRCFAR RDLVLRHQQK LHAALVGTGD PRRMTPAPNS TSSFASKRRH SVAADDPTDL 240
241 HIIKIAGNKE TILPTPKNLA GKTSEELKEA VVALAKSNNV ELPVSAPVMN DKREKTPPSK 300
301 AGSLGFREFK FSTKGVPVHS ASSDAVIDRA NTPSSMHKTK RHASFSASSA MTYMSSSNSP 360
361 HHSITNFELV EDAPHQVGFS TPQMTAKQLM ESVSELDLPP LTLDEPPQAI KFNLNLFNND 420
421 PSGQQQQQQQ QQQNSTSSTI VNSNNGSTVA TPGVYLLSSG PSLTDLLTMN SAHAGAGGYM 480
481 SSHHSPFDLG CFSHDKPTVS EFNLPSSFPN TIPSNSTTAS NSYSNLANQT YRQMSNEQPL 540
541 MSLSPKNPPT TVSDSSSTIN FNPGTNNLLE PSMEPNDKDS NIDPAAIDDK WLSEFINNSD 600
601 PKSTFKINFN HFNDIGFIYS PPSSRSSIPN KSPPNHSATS LNHEKASLSP RLNLSLNGST 660
661 DLPSTPQNQL KEPSYSDPIS HSSHKRRRDS VMMDYDLSNF FSSRQLDISK VLNGTEQNNS 720
721 HVNDDVLTLS FPGETDSNAT QKQLPVLTPS DLLSPFSVPS VSQVLFTNEL RSMMLADNNI 780
781 DSGAFPTTSQ LNDYVTYYKE EFHPFFSFIH LPSIIPNMDS YPLLLSISMV GALYGFHSTH 840
841 AKVLANAAST QIRKSLKVSE KNPETTELWV IQTLVLLTFY CIFNKNTAVI KGMHGQLTTI 900
901 IRLLKASRLN LPLESLCQPP IESDHIMEYE NSPHMFSKIR EQYNAPNQMN KNYQYFVLAQ 960
961 SRIRTCHAVL LISNLFSSLV GADCCFHSVD LKCGVPCYKE ELYQCRNSDE WSDLLCQYKI1020
1021 TLDSKFSLIE LSNGNEAYEN CLRFLSTGDS FFYGNARVSL STCLSLLISI HEKILIERNN1080
1081 ARISNNNTNS NNIELDDIEW KMTSRQRIDT MLKYWENLYL KNGGILTPTE NSMSTINANP1140
1141 AMRLIIPVYL FAKMRRCLDL AHVIEKIWLK DWSNMNKALE EVCYDMGSLR EATEYALNMV1200
1201 DAWTSFFTYI KQGKRRIFNT PVFATTCMFT AVLVISEYMK CVEDWARGYN ANNPNSALLD1260
1261 FSDRVLWLKA ERILRRLQMN LIPKECDVLK SYTDFLRWQD KDALDLSALN EEQAQRAMDP1320
1321 NTDINETIQL IVAASLSSKC LYLGVQILGD APIWPIILSF AHGLQSRAIY SVTKKRNTRI1380
1381 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
ELP2 ELP3 IKI3 NIS1 RSF2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
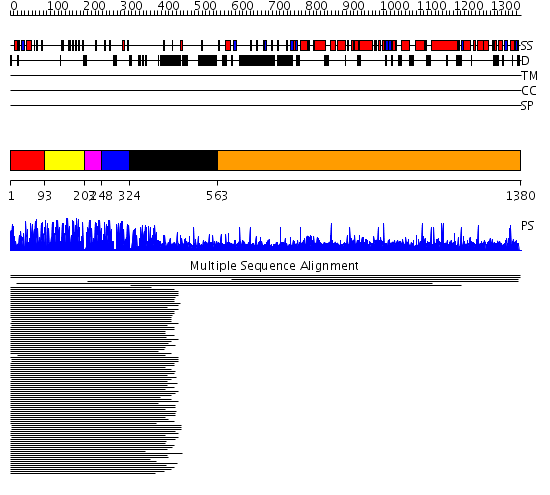
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | 61.144602 | Five-finger GLI1 | |
| 2 | View Details | [93..202] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [203..247] | 2.39794 | Tramtrack protein (two zinc-finger peptide) | |
| 4 | View Details | [248..323] | 4.824996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [324..562] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [563..1380] | 1.049001 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1218..1380] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|