













| General Information: |
|
| Name(s) found: |
TAH11 /
YJR046W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 604 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA pre-replicative complex [IMP cytoplasm [IDA |
| Biological Process: |
regulation of DNA replication initiation
[IDA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGTANSRRK EVLRVPVIDL NRVSDEEQLL PVVRAILLQH DTFLLKNYAN KAVLDALLAG 60
61 LTTKDLPDTS QGFDANFTGT LPLEDDVWLE QYIFDTDPQL RFDRKCRNES LCSIYSRLFK 120
121 LGLFFAQLCV KSVVSSAELQ DCISTSHYAT KLTRYFNDNG STHDGADAGA TVLPTGDDFQ 180
181 YLFERDYVTF LPTGVLTIFP CAKAIRYKPS TMATTDNSWV SIDEPDCLLF HTGTLLARWS 240
241 QGMHTTSPLQ IDPRANIVSL TIWPPLTTPI SSKGEGTIAN HLLEQQIKAF PKVAQQYYPR 300
301 ELSILRLQDA MKFVKELFTV CETVLSLNAL SRSTGVPPEL HVLLPQISSM MKRKIVQDDI 360
361 LKLLTIWSDA YVVELNSRGE LTMNLPKRDN LTTLTNKSRT LAFVERAESW YQQVIASKDE 420
421 IMTDVPAFKI NKRRSSSNSK TVLSSKVQTK SSNANALNNS RYLANSKENF MYKEKMPDSQ 480
481 ANLMDRLRER ERRSAALLSQ RQKRYQQFLA MKMTQVFDIL FSLTRGQPYT ETYLSSLIVD 540
541 SLQDSNNPIG TKEASEILAG LQGILPMDIS VHQVDGGLKV YRWNSLDKNR FSKLLQIHKS 600
601 KQQD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
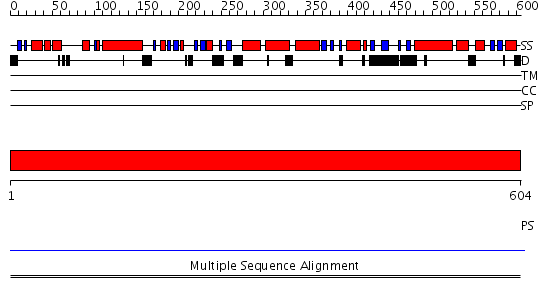
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..604] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [332..457] | 1.44 | Strucure of Geminin-Cdt1 complex | |
| 3 | View Details | [458..604] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)