













| General Information: |
|
| Name(s) found: |
YAP1801 /
YHR161C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 637 amino acids |
Gene Ontology: |
|
| Cellular Component: |
actin cortical patch
[IDA |
| Biological Process: |
endocytosis
[IPI |
| Molecular Function: |
clathrin binding
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTYFKLVKG ATKIKSAPPK QKYLDPILLG TSNEEDFYEI VKGLDSRIND TAWTIVYKSL 60
61 LVVHLMIREG SKDVALRYYS RNLEFFDIEN IRGSNGSASG DMRALDRYDN YLKVRCREFG 120
121 KIKKDYVRDG YRTLKLNSGN YGSSRNKQHS INIALDHVES LEVQIQALIK NKYTQYDLSN 180
181 ELIIFGFKLL IQDLLALYNA LNEGIITLLE SFFELSHHNA ERTLDLYKTF VDLTEHVVRY 240
241 LKSGKTAGLK IPVIKHITTK LVRSLEEHLI EDDKTHNTFV PVDSSQGSAG AVVAKSTAQE 300
301 RLEQIREQKR ILEAQLKNEQ VAISPALTTV TAAQSYNPFG TDSSMHTNIP MAVANQTQQI 360
361 ANNPFVSQTQ PQVMNTPTAH TEPANLNVPE YAAVQHTVNF NPVQDAGVSA QQTGYYSINN 420
421 HLTPTFTGAG FGGYSVSQDT TAASNQQVSH SQTGSNNPFA LHNAATIATG NPAHENVLNN 480
481 PFSRPNFDEQ NTNMPLQQQI ISNPFQNQTY NQQQFQQQKM PLSSINSVMT TPTSMQGSMN 540
541 IPQRFDKMEF QAHYTQNHLQ QQQQQQQQQQ QQQQQQPQQG YYVPATAGAN PVTNITGTVQ 600
601 PQNFPFYPQQ QPQPEQSQTQ QPVLGNQYAN NLNLIDM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CWP2 DBP2 FLC2 PPN1 RPS31 SRP40 TFA1 TFA2 YAP1801 |
| View Details | Qiu et al. (2008) |
|
ENT1 ENT2 ENT4 RVS167 YAP1801 YAP1802 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
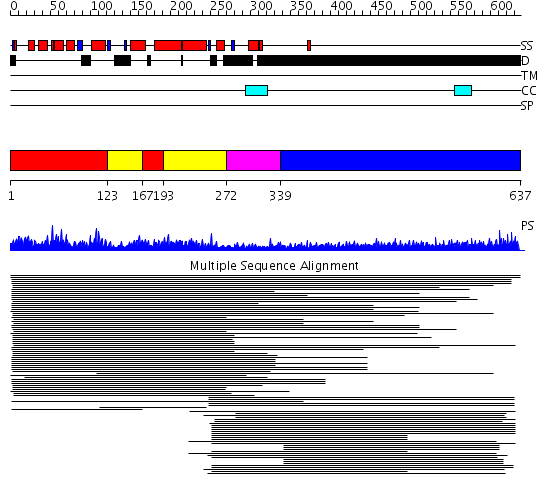
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] [167..192] |
298.519517 | Clathrin assembly lymphoid myeloid leukaemia protein, Calm | |
| 2 | View Details | [123..166] [193..271] |
298.519517 | Clathrin assembly lymphoid myeloid leukaemia protein, Calm | |
| 3 | View Details | [272..338] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [339..637] | 43.388997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)