













| General Information: |
|
| Name(s) found: |
POX1 /
YGL205W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 748 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisomal matrix
[TAS]
|
| Biological Process: |
fatty acid beta-oxidation
[TAS]
|
| Molecular Function: |
acyl-CoA oxidase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTRRTTINPD SVVLNPQKFI QKERADSKIK VDQVNTFLES SPERRTLTHA LIDQIVNDPI 60
61 LKTDTDYYDA KKMQEREITA KKIARLASYM EHDIKTVRKH FRDTDLMKEL QANDPDKASP 120
121 LTNKDLFIFD KRLSLVANID PQLGTRVGVH LGLFGNCIKG NGTDEQIRYW LQERGATLMK 180
181 GIYGCFAMTE LGHGSNVAQL QTRAVYDKQN DTFVIDTPDL TATKWWIGGA AHSATHAAVY 240
241 ARLIVEGKDY GVKTFVVPLR DPSTFQLLAG VSIGDIGAKM GRDGIDNGWI QFRNVVIPRE 300
301 FMLSRFTKVV RSPDGSVTVK TEPQLDQISG YSALLSGRVN MVMDSFRFGS KFATIAVRYA 360
361 VGRQQFAPRK GLSETQLIDY PLHQYRVLPQ LCVPYLVSPV AFKLMDNYYS TLDELYNASS 420
421 SAYKAALVTV SKKLKNLFID SASLKATNTW LIATLIDELR QTCGGHGYSQ YNGFGKGYDD 480
481 WVVQCTWEGD NNVLSLTSAK SILKKFIDSA TKGRFDNTLD VDSFSYLKPQ YIGSVVSGEI 540
541 KSGLKELGDY TEIWSITLIK LLAHIGTLVE KSRSIDSVSK LLVLVSKFHA LRCMLKTYYD 600
601 KLNSRDSHIS DEITKESMWN VYKLFSLYFI DKHSGEFQQF KIFTPDQISK VVQPQLLALL 660
661 PIVRKDCIGL TDSFELPDAM LNSPIGYFDG DIYHNYFNEV CRNNPVEADG AGKPSYHALL 720
721 SSMLGRGFEF DQKLGGAANA EILSKINK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
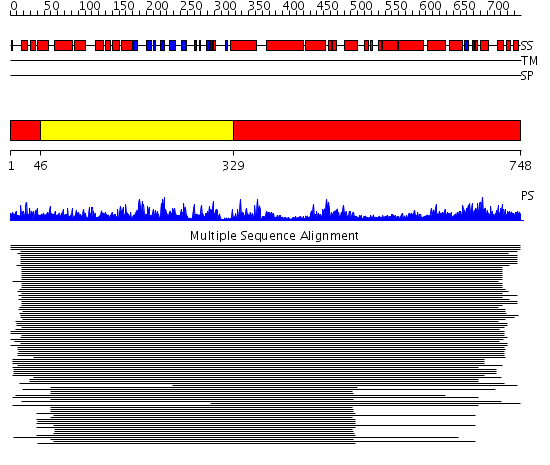
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..45] [329..748] |
917.228787 | Peroxisomal acyl-CoA oxidase-II, domains 3 and 4; Peroxisomal acyl-CoA oxidase-II, domains 1 and 2 | |
| 2 | View Details | [46..328] | 917.228787 | Peroxisomal acyl-CoA oxidase-II, domains 3 and 4; Peroxisomal acyl-CoA oxidase-II, domains 1 and 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)