













| General Information: |
|
| Name(s) found: |
DIN7 /
YDR263C
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 430 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
DNA repair
[IDA |
| Molecular Function: |
nuclease activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIPGLLPQL KRIQKQVSLK KYMYQTLAID GYAWLHRASC ACAFELVMNK PTNKYLQFFI 60
61 KRLQLLKRLK IKPYIVFDGD SLFVKNHTET RRRKKRLENE MIAKKLWSAG NRYNAMEYFQ 120
121 KSVDITPEMA KCIIDYCKLH SIPYIVAPFE ADPQMVYLEK MGLIQGIISE DSDLLVFGCK 180
181 TLITKLNDQG KALEISKDDF SALPENFPLG ELSEQQFRNL VCLAGCDYTS GIWKVGVVTA 240
241 MKIVKRYSEM KDILIQIERT EKLCFSKAFK QQVEFANYAF QYQRVFCPLS NQITTLNNIP 300
301 KAVTNSHAEI IKIMKCIGSV VERGSGVRKD VINTKNIDHK VHEMIAKGEL HPVDMASKLI 360
361 NRERKLKARK LFKVGLLGGE SNSFNKKVEQ PLVDTQDVLS ERENSLDNKN ASSIYMTSPA 420
421 AISGTVPSIF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
DIN7 TDA1 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
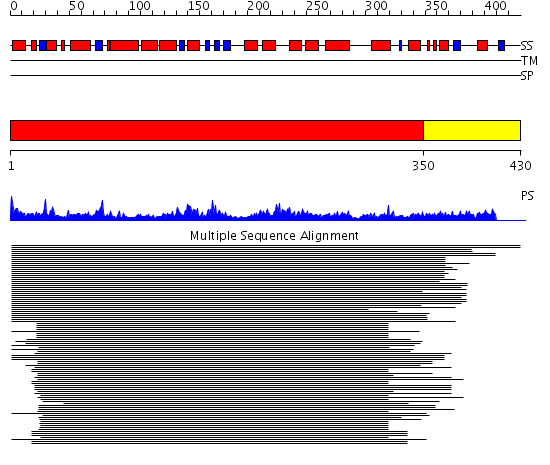
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..349] | 164.0103 | Flap endonuclease-1 (Fen-1 nuclease) | |
| 2 | View Details | [350..430] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)