













| General Information: |
|
| Name(s) found: |
TDA1 /
YMR291W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 586 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
protein kinase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTASSSASQ LQQRLPEEKP WPQLSGSNAD AQTFKCKYVT NHNSLGDGNF SVVKECMNIH 60
61 TKDLYAMKLI KKQTVKNKIQ LIQREFDLLR SISEKIRDME KKNEHSLDIF EGHHHILQLF 120
121 DYFETADNIV LITQLCQKGD LYEKIVENQC LDLETQVTSY CACLVSVLEF LHSQGIVHRD 180
181 LKAENVLFRL RVNENEKNLQ GEHHGDFKYD LLAHDLVLAD FGLAAEYNTS KVNSLKEFVG 240
241 TISYIAPEIV KCKGVGEMTP DQVGKLDKYG CPVDIWALGV LTYFMAFGYT PFDCTTDDET 300
301 LECISKCDYY VDEQMMHDPK YEQFWNFVQC CFTIDPAVRR SAKNLKQHPF IKDYFATSNS 360
361 LNTKDTPNFS FHPTIRRVSS TASMHTLRSP SKSRKTTTLA YLNMDGGSSE TSTAFSSKMD 420
421 LPDLYVDRTI NSRERSLNRI RDTLKKTLSM TSLKPAGTFD YLHANKNGTS LSSSKSGLVK 480
481 KNSTFVLDPK PPKNSLMNGC FSTTPESRSN FNTPKTLSRQ GSSTSVKKYV NEVDLLLTPR 540
541 TASMSSNDTT AINDYDTTND KNPARKHAAS FQVNVDDSDG DETMQI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
DIN7 TDA1 |
| View Details | Ho Y, et al. (2002) |
|
FUM1 TDA1 VPS33 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
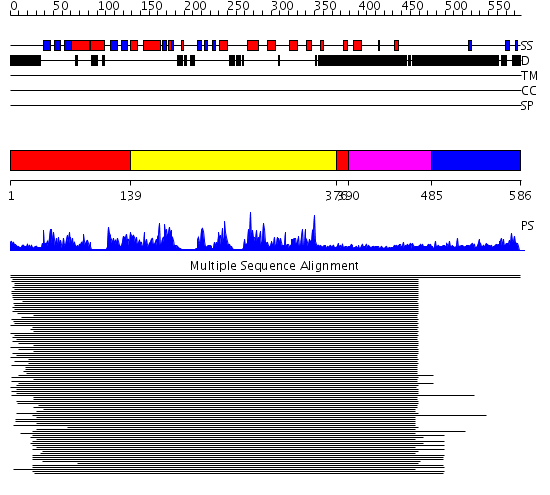
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] [376..389] |
330.228787 | Calmodulin-dependent protein kinase | |
| 2 | View Details | [139..375] | 330.228787 | Calmodulin-dependent protein kinase | |
| 3 | View Details | [390..484] | 3.30103 | No description for 1ew7C_ was found. | |
| 4 | View Details | [485..586] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)