













| General Information: |
|
| Name(s) found: |
gi|11559516
[NCBI NR]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Plasmodium falciparum |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: |
pre-replicative complex
[ISS |
| Biological Process: |
DNA strand elongation involved in DNA replication
[TAS |
| Molecular Function: |
ATP-dependent DNA helicase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIGIQEGRAF FNPTRNPNNR PENNIRIEEN LPSLNEVYNY FQDFLSRYSS NTLKEGNLKR 60
61 TLFENVKNNN FTLDLKLDDI YKQQTVFENN EISATPRSER KMKKPENYSA ILARALSERP 120
121 LTYLPTIERV CYEVCETANI LNDEDEHLNY IQINLLNTFI RPTPIRGLLA ATQERFVVVP 180
181 GIIVQASKPQ HKMRKITLQC RYCDHKMSID VPLWRDKPQL PPYCRYSSTM KSSMGLGNAM 240
241 DSQLGCNGVL EPYVILPNEC TFVDIQSLKM QELPEAVPTG DMPRHLQLNV TRYLCEKMIP 300
301 GDRVYVHGVL TSYNPNPRPT RADGTNFSYL HVLGFQKYDD MSGNDLNFDV EERNELTLLA 360
361 AEHNIHEKIF KSIAPELYGM DEVKKACACL LFGGTRKRIG EETKIRGDIN MLMLGDPSVA 420
421 KSQILKFVNR CAPVSVYTSG KGSSAAGLTA AVMRDSQGVF SLEGGAMVLA DGGVVCIDEF 480
481 DKMRDDDVVA IHEAMEQQTI SISKAGITTM LNTRCSVIAA ANPSFGSYDD SQDTTYQHDF 540
541 KTTILSRFDI IFLLRNKQDV EKDTLLCNHI VALHASKHKS QEGEIPLSKL TRYIQYAKRE 600
601 IAPLLSKEAR DSLRNFYVQT RAEYRGDRRS VTKKIPITLR QLESLIRLAE SFAKMELSQF 660
661 ATDKHVQMSI DLFSASTAET AKQCLIFETM SPLEQKAVKQ AEDAILGRLG KGQRASRVNL 720
721 FRELQLRGFD RSALSKALAI LIKKGELQER GDRSVRRT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
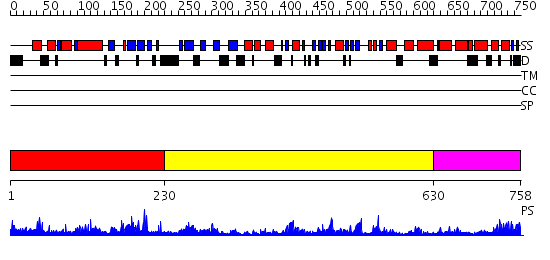
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..229] | 37.39794 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 2 | View Details | [230..629] | 33.0 | Crystal Structure Analysis of ClpB | |
| 3 | View Details | [630..758] | 48.69897 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)