













| General Information: |
|
| Name(s) found: |
gi|29653825
[NCBI NR]
gi|29541088 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Coxiella burnetii RSA 493 |
| Length: | 269 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
amino acid transport
[ISS |
| Molecular Function: |
ATPase activity, coupled to transmembrane movement of substances
[ISS binding [ISS amino acid-transporting ATPase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKKNNNRAVS RFFRWIPAFV RMTGTFYFVS CLFVSPVFAT DTIKFATEAT YPPYVYMGPS 60
61 GQVEGFGADI VKAVCKQMQA VCTISNQPWD SLIPSLKLGK FDALFGGMNI TTARQKEVDF 120
121 TDPYYTNSVS FIADKNTPLT LSKQGLKGKI IGVQGGTTFD SYLQDSFGNS ITIQRYPSEE 180
181 DALMDLTSGR VDAVVGDTPL IKQWLKQNGR REYVLIGKPV NDPNYFGKGV GIAVKKGNQA 240
241 LLLKLNKALA AIKANGVYAA IVQKYFGQE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
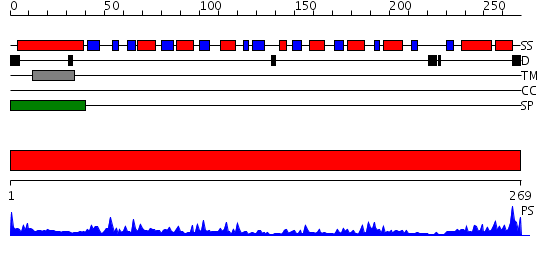
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..269] | 52.09691 | Histidine-binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|