













| General Information: |
|
| Name(s) found: |
gi|24375187
[NCBI NR]
gi|24349970 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 475 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
two-component signal transduction system (phosphorelay)
[ISS cellular response to nitrogen starvation [ISS |
| Molecular Function: |
two-component response regulator activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTILIVDDN HAICQALGLM LEINGYQVLT CHTPDDALNL IAAHDIDLVI QDMNFTRDTT 60
61 SGEEGRQLFY ALRERQPDLP IILMTAWTQL ETAVELVKVG AADYMGKPWD DAKLLNSITN 120
121 LVALYKLACT NHQLERVNSQ RQVAIADAEL CGIVFGSGAM QRCIDLALQL ARSDVSVLIT 180
181 GPNGAGKDKL ADIIHANSPL KNKPFIKVNV GALPMELLEA ELFGAEAGAF TGANKARMGR 240
241 FEAADGGTLF LDEIGNLPLS GQVKLLRVLQ TGEFERLGSH KTLKVKVRVI SATNADLAQD 300
301 IAEGRFREDL FYRLNVIELA LPPLNQRIDD ILPLVKHFVG EGFSLSKPAQ QALLQHRWPG 360
361 NVRELENACK RAALLAQSRV LTEADFGLPP VQARQLSATN AFALGQAAHL AAAFEPQSHQ 420
421 SVFYSEIHSE AQDVSREDIE AALQQHNGVI ARVAKSLGLS RQALYRRMDK FGLEK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
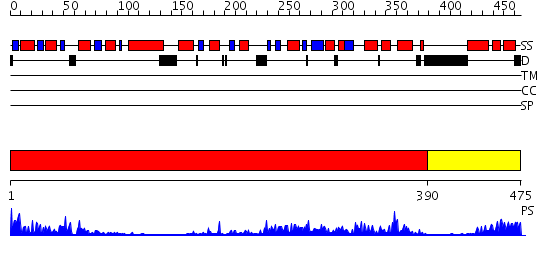
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..389] | 74.30103 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [390..475] | 68.69897 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)