













| General Information: |
|
| Name(s) found: |
VPS34_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 875 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLNNITFCV SQDLDVPLKV KIKSLEGHKP LLKPSQKILN PELMLIGSNV FPSSDLIVSL 60
61 QVFDKERNRN LTLPIYTPYI PFRNSRTWDY WLTLPIRIKQ LTFSSHLRII LWEYNGSKQI 120
121 PFFNLETSIF NLKDCTLKRG FESLKFRYDV IDHCEVVTDN KDQENLNKYF QGEFTRLPWL 180
181 DEITISKLRK QRENRTWPQG TFVLNLEFPM LELPVVFIER EIMNTQMNIP TLKNNPGLST 240
241 DLREPNRNDP QIKISLGDKY HSTLKFYDPD QPNNDPIEEK YRRLERASKN ANLDKQVKPD 300
301 IKKRDYLNKI INYPPGTKLT AHEKGSIWKY RYYLMNNKKA LTKLLQSTNL REESERVEVL 360
361 ELMDSWAEID IDDALELLGS TFKNLSVRSY AVNRLKKASD KELELYLLQL VEAVCFENLS 420
421 TFSDKSNSEF TIVDAVSSQK LSGDSMLLST SHANQKLLKS ISSESETSGT ESLPIVISPL 480
481 AEFLIRRALV NPRLGSFFYW YLKSESEDKP YLDQILSSFW SRLDKKSRNI LNDQVRLINV 540
541 LRECCETIKR LKDTTAKKME LLVHLLETKV RPLVKVRPIA LPLDPDVLIC DVCPETSKVF 600
601 KSSLSPLKIT FKTTLNQPYH LMFKVGDDLR QDQLVVQIIS LMNELLKNEN VDLKLTPYKI 660
661 LATGPQEGAI EFIPNDTLAS ILSKYHGILG YLKLHYPDEN ATLGVQGWVL DNFVKSCAGY 720
721 CVITYILGVG DRHLDNLLVT PDGHFFHADF GYILGQDPKP FPPLMKLPPQ IIEAFGGAES 780
781 SNYDKFRSYC FVAYSILRRN AGLILNLFEL MKTSNIPDIR IDPNGAILRV RERFNLNMSE 840
841 EDATVHFQNL INDSVNALLP IVIDHLHNLA QYWRT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
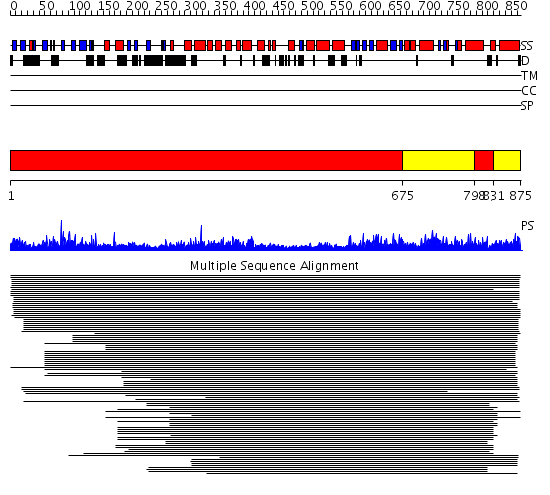
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..674] [798..830] |
666.228787 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 2 | View Details | [675..797] [831..875] |
666.228787 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)