













| General Information: |
|
| Name(s) found: |
clpB /
EG10157
[EcoGene]
|
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli str. K-12 substr. MG1655 |
| Length: | 857 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane fraction
[IDA membrane [IDA cytosol [IDA |
| Biological Process: |
protein metabolic process
[IEA]
response to stress [IEA] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA] protein binding [IEA] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLDRLTNKF QLALADAQSL ALGHDNQFIE PLHLMSALLN QEGGSVSPLL TSAGINAGQL 60
61 RTDINQALNR LPQVEGTGGD VQPSQDLVRV LNLCDKLAQK RGDNFISSEL FVLAALESRG 120
121 TLADILKAAG ATTANITQAI EQMRGGESVN DQGAEDQRQA LKKYTIDLTE RAEQGKLDPV 180
181 IGRDEEIRRT IQVLQRRTKN NPVLIGEPGV GKTAIVEGLA QRIINGEVPE GLKGRRVLAL 240
241 DMGALVAGAK YRGEFEERLK GVLNDLAKQE GNVILFIDEL HTMVGAGKAD GAMDAGNMLK 300
301 PALARGELHC VGATTLDEYR QYIEKDAALE RRFQKVFVAE PSVEDTIAIL RGLKERYELH 360
361 HHVQITDPAI VAAATLSHRY IADRQLPDKA IDLIDEAASS IRMQIDSKPE ELDRLDRRII 420
421 QLKLEQQALM KESDEASKKR LDMLNEELSD KERQYSELEE EWKAEKASLS GTQTIKAELE 480
481 QAKIAIEQAR RVGDLARMSE LQYGKIPELE KQLEAATQLE GKTMRLLRNK VTDAEIAEVL 540
541 ARWTGIPVSR MMESEREKLL RMEQELHHRV IGQNEAVDAV SNAIRRSRAG LADPNRPIGS 600
601 FLFLGPTGVG KTELCKALAN FMFDSDEAMV RIDMSEFMEK HSVSRLVGAP PGYVGYEEGG 660
661 YLTEAVRRRP YSVILLDEVE KAHPDVFNIL LQVLDDGRLT DGQGRTVDFR NTVVIMTSNL 720
721 GSDLIQERFG ELDYAHMKEL VLGVVSHNFR PEFINRIDEV VVFHPLGEQH IASIAQIQLK 780
781 RLYKRLEERG YEIHISDEAL KLLSENGYDP VYGARPLKRA IQQQIENPLA QQILSGELVP 840
841 GKVIRLEVNE DRIVAVQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
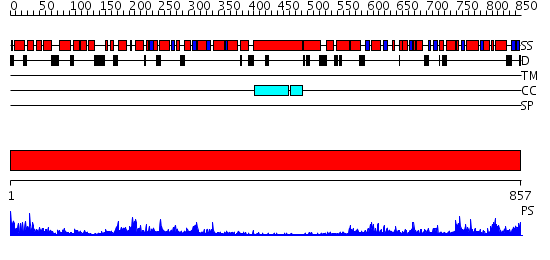
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..857] | 171.0 | Crystal Structure Analysis of ClpB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)