













| General Information: |
|
| Name(s) found: |
RAD51_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 342 amino acids |
Gene Ontology: |
|
| Cellular Component: |
[NOT]nucleus
[IEP]
|
| Biological Process: |
DNA repair
[ISS]
response to radiation [NAS] DNA metabolic process [IDA][TAS] response to gamma radiation [IEP] double-strand break repair [IMP] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA] DNA binding [IEA] DNA-dependent ATPase activity [IEA] protein binding [IEA] nucleoside-triphosphatase activity [IEA] sequence-specific DNA binding [IEA] damaged DNA binding [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTMEQRRNQ NAVQQQDDEE TQHGPFPVEQ LQAAGIASVD VKKLRDAGLC TVEGVAYTPR 60
61 KDLLQIKGIS DAKVDKIVEA ASKLVPLGFT SASQLHAQRQ EIIQITSGSR ELDKVLEGGI 120
121 ETGSITELYG EFRSGKTQLC HTLCVTCQLP MDQGGGEGKA MYIDAEGTFR PQRLLQIADR 180
181 FGLNGADVLE NVAYARAYNT DHQSRLLLEA ASMMIETRFA LLIVDSATAL YRTDFSGRGE 240
241 LSARQMHLAK FLRSLQKLAD EFGVAVVITN QVVAQVDGSA LFAGPQFKPI GGNIMAHATT 300
301 TRLALRKGRA EERICKVISS PCLPEAEARF QISTEGVTDC KD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
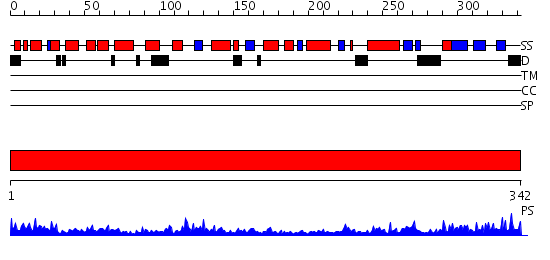
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..342] | 74.522879 | A Crystal Structure of the Rad51 Filament |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)