













| General Information: |
|
| Name(s) found: |
VPS5 /
YOR069W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 675 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA retromer complex [IPI cytosol [IDA |
| Biological Process: |
protein retention in Golgi apparatus
[IMP retrograde transport, endosome to Golgi [IPI |
| Molecular Function: |
protein transporter activity
[IMP phosphatidylinositol-3-phosphate binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDYEDNLEAP VWDELNHEGD KTQSLVSNSI ESIGQISTNE ERKDNELLET TASFADKIDL 60
61 DSAPEWKDPG LSVAGNPQLE EHDNSKADDL INSLAPEQDP IADLKNSTTQ FIATRESGGA 120
121 LFTGNANSPL VFDDTIYDAN TSPNTSKSIS GRRSGKPRIL FDSARAQRNS KRNHSLKAKR 180
181 TTASDDTIKT PFTDPLKKAE KENEFVEEPL DDRNERRENN EGKFTASVEK NILEQVDRPL 240
241 YNLPQTGANI SSPAEVEENS EKFGKTKIGS KVPPTEKAVA FKVEVKDPVK VGELTSIHVE 300
301 YTVISESSLL ELKYAQVSRR YRDFRWLYRQ LQNNHWGKVI PPPPEKQSVG SFKENFIENR 360
361 RFQMESMLKK ICQDPVLQKD KDFLLFLTSD DFSSESKKRA FLTGSGAIND SNDLSEVRIS 420
421 EIQLLGAEDA AEVLKNGGID AESHKGFMSI SFSSLPKYNE ADEFFIEKKQ KIDELEDNLK 480
481 KLSKSLEMVD TSRNTLAAST EEFSSMVETL ASLNVSEPNS ELLNNFADVH KSIKSSLERS 540
541 SLQETLTMGV MLDDYIRSLA SVKAIFNQRS KLGYFLVVIE NDMNKKHSQL GKLGQNIHSE 600
601 KFREMRKEFQ TLERRYNLTK KQWQAVGDKI KDEFQGFSTD KIREFRNGME ISLEAAIESQ 660
661 KECIELWETF YQTNL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
MUK1 PEP8 VPS17 VPS29 VPS35 VPS5 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
VPS17 VPS5 |
| View Details | Krogan NJ, et al. (2006) |

|
MIA40 MUK1 PEP8 VPS17 VPS29 VPS35 VPS5 |
| View Details | Gavin AC, et al. (2006) |
|
VPS29 VPS35 VPS5 |
| View Details | Gavin AC, et al. (2002) |

|
CLU1 CRN1 ECM29 KAP123 MEC1 MIP6 MIS1 MRPL23 NAB3 NAB6 NRD1 ROM2 RPA135 SEN54 SUI3 SUP35 SUP45 TAO3 USO1 UTP20 VAC14 VPS5 YJL017W |
| View Details | Qiu et al. (2008) |

|
ATG20 PEP8 SNX41 VPS17 VPS29 VPS35 VPS5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #27 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps3-5 | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
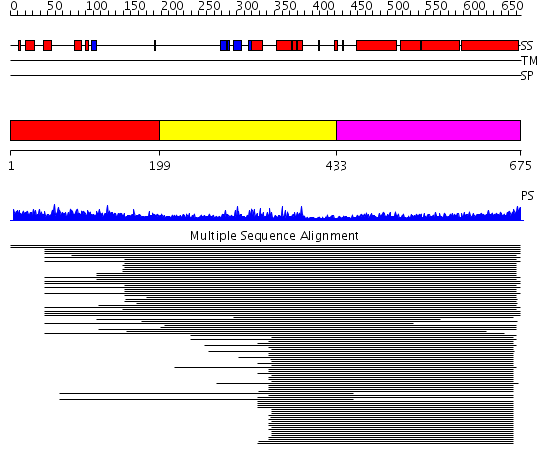
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..198] | 33.0 | Kinesin | |
| 2 | View Details | [199..432] | 35.522879 | Heavy meromyosin subfragment | |
| 3 | View Details | [433..675] | 35.522879 | Heavy meromyosin subfragment | |
| 4 | View Details | [345..675] | 41.0 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)