













| General Information: |
|
| Name(s) found: |
JLP1 /
YLL057C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 412 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
sulfur metabolic process
[IDA |
| Molecular Function: |
sulfonate dioxygenase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPAAAQTAI PLPSTDLPVK IITNGLKNLN YTSKQGYGNF DTHFYDGQDE VSPSGLLKIR 60
61 KSYREKSKYP DYLPTWDPTE KYGPLEFHEY HDPALRADGN FSNLFAKENV GQLKVKKITP 120
121 KLGLEINGIQ LTDLSDAAKD ELALLVAQKG VVVFRNQNFA DEGPDYVTEY GRHFGKLHIH 180
181 QTSGHPQNNP ELHITFRRPD AEEFARVFDD STSSGGWHTD VSYELQPPSY TFFSVVEGPD 240
241 GGGDTLFADT IEAFDRLSKP LQDFLSTLHV IHSSKEQAEN SQRQGGIKRR APVTHIHPLV 300
301 RVHPVLKKKC LYVNRAFSRK IVELKRQESE SLLNFLYNLV ESSHDLQLRA KWEPHSVVIW 360
361 DNRRVQHSAV IDWEEPIHRH AFRITPQAER PVEDLKFLND ENYYPSSLTL DI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
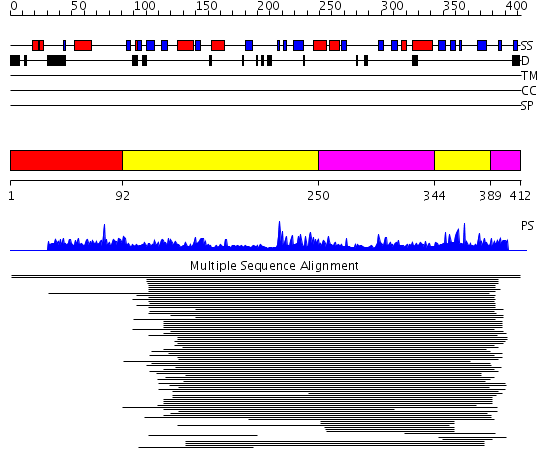
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [92..249] [344..388] |
386.522879 | Taurine/alpha-ketoglutarate dioxygenase | |
| 3 | View Details | [250..343] [389..412] |
386.522879 | Taurine/alpha-ketoglutarate dioxygenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)