













| General Information: |
|
| Name(s) found: |
ATG11 /
YPR049C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1178 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extrinsic to membrane
[IDA |
| Biological Process: |
peroxisome degradation
[IMP telomere maintenance [IMP protein targeting to vacuole [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADADEYSTA PTQQEITPLQ TTATIINAIS GECITTNVDF FVSLDKFKQF IARKWKIPPD 60
61 QLLILLPYGN KLKPSMFKEL LINRSFTLND FYVYDRRLFS LVSKPTPTNL LTSKDSNPMN 120
121 SPNSNDLTET LEYLIKNSHI SQYQGSDTIM IKPMPSPLED ADVDLSRLNY HSVTSLLTTN 180
181 LGWLSALEID VHYFKSLIPD IIAHIKRIFD GLTVCSQYLK LYCFDVESLY NSNVQFLNQL 240
241 VDNGMTSKWE KCFNDTLSKL TALEGDSLQK FINIESLLEN EKSVKILNHS INGKLNKIKR 300
301 EIDENASFRD IITVNIDRLR QMFTPNESKF ELEDQMAESF EVLVSEMRTR SRNVLDKEEE 360
361 EFNSQEFLKS MNVMLEKDKK ESVKTLFTIS QALYSQIGEL IDLKKSLQKH AVAILGNIAF 420
421 TQMEILGIKR LLLNECNKDL ELYKKYEVEF AQVEDLPLIY GLYLIEKYRR LSWFQQILSF 480
481 ISNFNQDLEL FKQNELRTRN KWVKNFGSIA TVFCEDLLSS SDFKRLNEYH SHTSPPNEDE 540
541 EDENENSIAN YRQDLVKVSQ AIDNYMTQIK ETDVSEPIID LLSKTLFETK RFHIIYSNFK 600
601 NNNNNSSNGN SISPEGSIAL KSDDVVKGYK TRIKKLESLL HEFQYSDIGH WPQGVLNTHL 660
661 KPFRGSATSI NKKKFLGASV LLEPANISEV NIDSVSQANN HQIQELESNV DDLLHQLQLL 720
721 KEENNRKSMQ ISEMGKKISD LEVEKTAYRE TLTNLNQELA RLTNEEQSHR TEIFTLNASF 780
781 KKQLNDIISQ DNEKIEKLTG DYDDVSKSRE RLQMDLDESN KKHEQEVNLL KADIERLGKQ 840
841 IVTSEKSYAE TNSSSMEKGE KFETIPLAED PGRENQISAY TQTLQDRIFD IISTNIFILE 900
901 NIGLLLTFDN NNNIQIRRVK GLKKGTAQSN ILDESTQMLD AHDNSLIKSP VFQKLKDEYE 960
961 LIKSVANGSE KDTQQSIFLG NITQLYDNKL YEVAVIRRFK DIETLAKKLT KENKIKRTLL1020
1021 ERFQREKVTL RNFQIGDLAL FLPTRENVNS VGSMSSSTSS LSSSFSSVDL STPPPLDAMS1080
1081 IQSSPSVIHS NVINQASISG RDKNKLMRPW AAFTAFEEST RYFLKDEKGL TKGKEWFVGR1140
1141 IVTLEHFVAD SPSNNPFRLP KGSVWFQVTA VVVSYQGV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
ATG11 ATG17 |
| View Details | Krogan NJ, et al. (2006) |
|
ATG11 ATG17 ATG29 CIS1 THI11 YBR197C |
| View Details | Gavin AC, et al. (2006) |
|
ATG11 ATG17 ATG29 |
| View Details | Ho Y, et al. (2002) |

|
AAC1 AAC3 ADE5,7 AHA1 ARC1 ARO1 ARP2 ATG11 ATG12 ATG17 CAR2 CPA2 CPR6 FET4 GCD11 GFA1 GUS1 IPP1 PPX1 PRB1 PTC7 REP1 REX2 RPN10 RPN11 RPN3 RPN5 RPN6 RPN7 RPT1 RPT3 SEC18 TCB1 TIF1, TIF2 TYR1 YHR020W YHR033W YNL208W |
| View Details | Qiu et al. (2008) |

|
ATG11 ATG2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
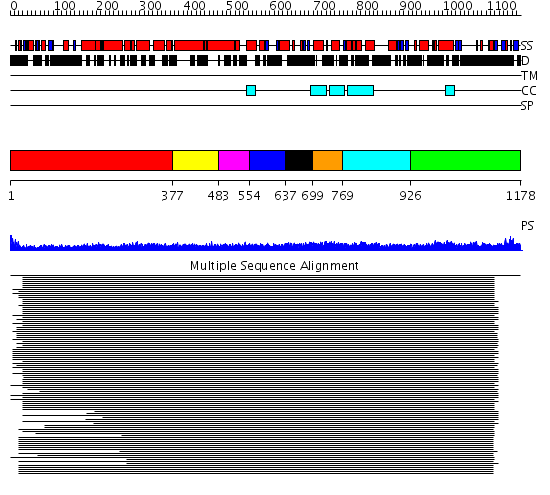
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..376] | 24.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [377..482] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [483..553] | 8.30103 | Tropomyosin | |
| 4 | View Details | [554..636] | 8.30103 | Tropomyosin | |
| 5 | View Details | [637..698] | 8.30103 | Tropomyosin | |
| 6 | View Details | [699..768] | 8.30103 | Tropomyosin | |
| 7 | View Details | [769..925] | 4.0 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 8 | View Details | [926..1178] | 27.829997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)