













| General Information: |
|
| Name(s) found: |
HIR2 /
YOR038C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 875 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA HIR complex [IDA |
| Biological Process: |
DNA replication-independent nucleosome assembly
[IDA regulation of transcription from RNA polymerase II promoter [IDA RNA elongation from RNA polymerase II promoter [IGI |
| Molecular Function: |
transcription corepressor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLLKYPLDI HNEQVNALAA LGPYIILAGS GGHVMAWRQQ QLVDTAFDRV MIKDLKPEVS 60
61 FQVDQDTTGD IFFITGDLET LYIGSEHRLW GYSGWLCRDT NNINSVEKMN SKLLFECKSP 120
121 STITDVKYDI NLGILFVLLS NENKILLFRH KTFDKLSEIT IDKASKPITG IIDPTGQTFT 180
181 VMTSDRSILV YQINKTGTHK LINKLTQHVQ MYPLHYRISM SPQADILPVI NSVKGVPNNA 240
241 TSCTALLDRN NNYKVTKTLV TPSSNGCRVL VYSPAFYEKP NLKKGTSTRY NLIATSGSTD 300
301 GTILVWNTKR MKPLFNALQV SSTAINDMSW SQDGFTLFAI SNDATLYTFA FQEKDLGVAL 360
361 PQTEIKSLQE VNKKLPKLEE PLAEQIPKSF PENIKLEESA SAAPIPNDIG RSAVGKKPTK 420
421 KKTANNQTNG IKTIQSTSME FNTPSYTVPR DLKRKPKEAT PSNIAPGSKK QKKELQPIDF 480
481 LDTGLLLPNT SFSRIRLATP KIRSTFKYSP INNPNLILDV KNGSGNEQRP TIVKLTSKVL 540
541 DQDQVLFQDF IPKLITICTA GDTFWSFCSE DGSIYIYSDS GRKLMAPLVL GVSISFLEAC 600
601 GTYLLCLTSI GELYCWNIEQ KKLAFPTNTI YPLLNPSLRY SDDILTRAEN ITLCSITKKG 660
661 VPLVTLSNGD GYLFDKNMET WLLVSDGWWA YGSQYWDTTN TTGLSSSKAN TDSFNGSESN 720
721 INEIVSDIKN DNQSIINFLE CKTNDELNRK GRIKNLQRFA RTILMKEGFE NMEEIVTLSH 780
781 LENKILISIR LEEPEEFSKL MMVYCIRLSE LGYMDRLNDV FQWLYDDLPI SGTGSAFADK 840
841 DFKRNLLKKI LIACGDIRQV QRVTTRYAKE MNIIS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
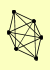
|
ASF1 HIR1 HIR2 HIR3 HPC2 RAD53 |
| View Details | Krogan NJ, et al. (2006) |
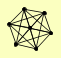
|
ASF1 HIR1 HIR2 HIR3 HPC2 QDR2 SFP1 |
| View Details | Ho Y, et al. (2002) |
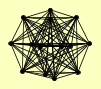
|
HHF1, HHF2 HIR2 HTA1 HTB1 KAP114 KRI1 NAP1 NOP4 NOP7 RET1 RPC82 RPO31 SPT16 UTP13 |
| View Details | Qiu et al. (2008) |
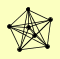
|
CAC2 HIR1 HIR2 HIR3 HPC2 MSI1 RLF2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | sample 3121 run on DecaLCQ: searched for phosphorylation | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | sample 3928 - Asf1-S-TEV-ZZ (in hir3delta mutant) | Green EM, et al (2005) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | 1.652995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [86..473] | 70.39794 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 3 | View Details | [474..698] | 23.515997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [699..752] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [753..875] | 7.63 | 5' to 3' exonuclease domain of DNA polymerase Taq; Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|