













| General Information: |
|
| Name(s) found: |
CAC2 /
YML102W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 468 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin assembly complex
[IDA chromosome, centromeric region [IDA |
| Biological Process: |
DNA repair
[IMP chromatin silencing [IMP nucleosome assembly [IMP |
| Molecular Function: |
transcription regulator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEASHLQIYW HDSQPVYSLT FQKNSANDKL FTAGGDNKVR IWKLNRDENG QNGGVRKIES 60
61 LDFLGSLTHH EQAINVIRFN SKGDVLASAG DDGQVLLWKQ EDPNTQQESV VRPFGMDAET 120
121 SEADENKEKW VVWKRLRGGS GATAAAEIYD LAWSPDNRNI VVACMDNSIR LFDVGAGMLV 180
181 CGQSDHGHYV QGVAWDPLNQ FILSQSADRS LHVYGVILSS AGVVTGLKLR SKIAKAELPC 240
241 PGDVLRTNYL FHNETLPSFF RRCSISPCGG LVVIPSGVYK VAGDEVANCV YVYTRSGILN 300
301 SAGGVKNRPA IRIPSLKKPA LMAAFSPVFY ETCQKSVLKL PYKLVFAIAT TNEVLVYDTD 360
361 VLEPLCVVGN IHYSPITDLA WSEDGSTLLI SSTDGFCSYV SIDTETQFGS RIEPPAMHAE 420
421 PLDTDESAVA AKNQREAGGI VNMLPVKKIP CNSSDSKKRR IHPTPVDL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
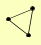
|
CAC2 MSI1 RLF2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
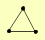
|
CAC2 MSI1 RLF2 |
| View Details | Krogan NJ, et al. (2006) |
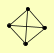
|
CAC2 FET5 MSI1 RLF2 |
| View Details | Gavin AC, et al. (2006) |

|
CAC2 MSI1 |
| View Details | Ho Y, et al. (2002) |

|
CAC2 RLF2 |
| View Details | Qiu et al. (2008) |
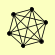
|
CAC2 HIR1 HIR2 HIR3 HPC2 MSI1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
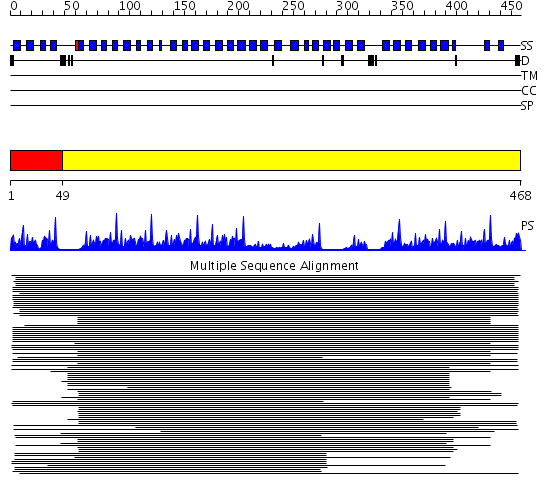
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..48] | 5.0 | WD domain, G-beta repeat No confident structure predictions are available. | |
| 2 | View Details | [49..468] | 78.491979 | Tup1, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)