













| General Information: |
|
| Name(s) found: |
CHS1 /
YNL192W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1131 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chitosome
[IDA plasma membrane [IDA |
| Biological Process: |
cytokinesis, completion of separation
[IMP cell budding [TAS |
| Molecular Function: |
chitin synthase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDQNNRSRN EYHSNRKNEP SYELQNAHSG LFHSSNEELT NRNQRYTNQN ASMGSFTPVQ 60
61 SLQFPEQSQQ TNMLYNGDDG NNNTINDNER DIYGGFVNHH RQRPPPATAE YNDVFNTNSQ 120
121 QLPSEHQYNN VPSYPLPSIN VIQTTPELIH NGSQTMATPI ERPFFNENDY YYNNRNSRTS 180
181 PSIASSSDGY ADQEARPILE QPNNNMNSGN IPQYHDQPFG YNNGYHGLQA KDYYDDPEGG 240
241 YIDQRGDDYQ INSYLGRNGE MVDPYDYENS LRHMTPMERR EYLHDDSRPV NDGKEELDSV 300
301 KSGYSHRDLG EYDKDDFSRD DEYDDLNTID KLQFQANGVP ASSSVSSIGS KESDIIVSND 360
361 NLTANRALKR SGTEIRKFKL WNGNFVFDSP ISKTLLDQYA TTTENANTLP NEFKFMRYQA 420
421 VTCEPNQLAE KNFTVRQLKY LTPRETELML VVTMYNEDHI LLGRTLKGIM DNVKYMVKKK 480
481 NSSTWGPDAW KKIVVCIISD GRSKINERSL ALLSSLGCYQ DGFAKDEINE KKVAMHVYEH 540
541 TTMINITNIS ESEVSLECNQ GTVPIQLLFC LKEQNQKKIN SHRWAFEGFA ELLRPNIVTL 600
601 LDAGTMPGKD SIYQLWREFR NPNVGGACGE IRTDLGKRFV KLLNPLVASQ NFEYKMSNIL 660
661 DKTTESNFGF ITVLPGAFSA YRFEAVRGQP LQKYFYGEIM ENEGFHFFSS NMYLAEDRIL 720
721 CFEVVTKKNC NWILKYCRSS YASTDVPERV PEFILQRRRW LNGSFFASVY SFCHFYRVWS 780
781 SGHNIGRKLL LTVEFFYLFF NTLISWFSLS SFFLFFRILT VSIALAYHSA FNVLSVIFLW 840
841 LYGICTLSTF ILSLGNKPKS TEKFYVLTCV IFAVMMIYMI FCSIFMSVKS FQNILKNDTI 900
901 SFEGLITTEA FRDIVISLGS TYCLYLISSI IYLQPWHMLT SFIQYILLSP SYINVLNIYA 960
961 FCNVHDLSWG TKGAMANPLG KINTTEDGTF KMEVLVSSSE IQANYDKYLK VLNDFDPKSE1020
1021 SRPTEPSYDE KKTGYYANVR SLVIIFWVIT NFIIVAVVLE TGGIADYIAM KSISTDDTLE1080
1081 TAKKAEIPLM TSKASIYFNV ILWLVALSAL IRFIGCSIYM IVRFFKKVTF R |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
CHS1 ENO2 PGK1 |
| View Details | Ho Y, et al. (2002) |
|
ARO1 BCK1 CHS1 CMP2 DBP7 KOG1 KSP1 PRI2 PSY2 TPD3 |
| View Details | Qiu et al. (2008) |
|
CHS1 CHS3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
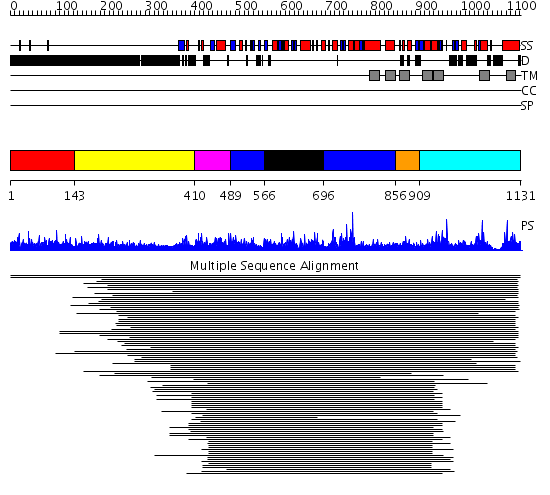
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 2.69897 | Fibrinogen | |
| 2 | View Details | [143..409] | 14.32 | RBP1 | |
| 3 | View Details | [410..488] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [489..565] [696..855] |
7.7 | Diol dehydratase, alpha subunit | |
| 5 | View Details | [566..695] | 7.7 | Diol dehydratase, alpha subunit | |
| 6 | View Details | [856..908] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [909..1131] | 1.122952 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|