













| General Information: |
|
| Name(s) found: |
NGL2 /
YMR285C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 515 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IDA |
| Biological Process: |
rRNA processing
[IMP |
| Molecular Function: |
endoribonuclease activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTQDKEVKVV APDVAPDQEV EINKSVKDAK HQTNDDSLLQ HKKKGKKGKK SKPIVTPEHI 60
61 AKVRAEREVM RKAKRDAMLA QGVDPDCPPE LHFIRRPFLS LHEAEPVTGF RFKLMTYNCL 120
121 AQALIRRKLF PDSGDALKWY RRSKVLLNEF KYYNSDVICL QEIDHIQFQS FWKDEFSKLG 180
181 YDGQYYRNAT KNHGVAIMWR RELFHQVDKM LIDYDKESSE SISTRTTTNN VGLVLALKFS 240
241 EKVLSNLGKK SSKKCGILIG TTHLFWHPFG TYERTRQCYI VLKKMKEFMH RVNVLQNEND 300
301 GDLSHWFPFF CGDFNSQPFD TPYLSMTSKP VHYRNRAKTV IGCSTSYKFS KVRDGEEGAD 360
361 DEEGGNIEKY GKDQPESPVP EKFHANEEQS ELVDKMAQLH NSLDMRAISL YSVGYKNVHP 420
421 ENAGLDNDRG EPEISNWANT WRGLLDYLFY VKKWDPQSNC QEVETLGDFE KENKVKCRGF 480
481 LRMPPGNEMT KHGQPHVGEY ASDHLSMVCD LELQL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
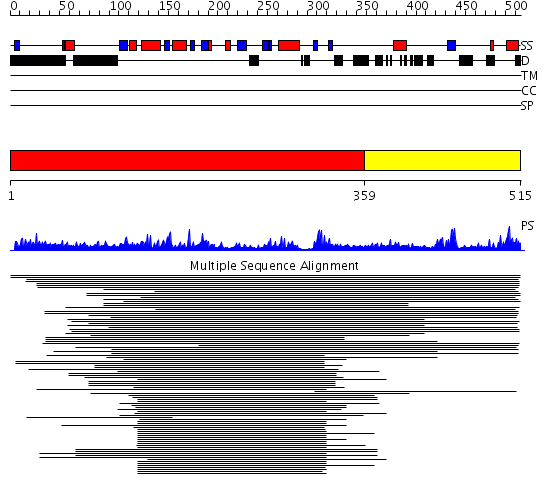
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..358] | 23.30103 | DNA repair endonuclease Hap1 | |
| 2 | View Details | [359..515] | 1.058983 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)