













| General Information: |
|
| Name(s) found: |
QNS1 /
YHR074W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 714 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
NAD biosynthetic process
[IDA |
| Molecular Function: |
NAD+ synthase (glutamine-hydrolyzing) activity
[IDA hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHLITLATC NLNQWALDFE GNRDRILQSI KIAKERGARL RVGPELEITG YGCLDHFLEN 60
61 DVCLHSWEMY AQIIKNKETH GLILDIGMPV LHKNVRYNCR LLSLDGEILF IRPKIWLAND 120
121 GNYREMRFFT PWMKPGVVED FILPPEIQKV TGQRLVPFGD AVINSLDTCI GTETCEELFT 180
181 PQSPHIAMSL DGVEIMTNSS GSHHELRKLN KRLDLILNAT KRCGGVYLYA NQRGCDGDRL 240
241 YYDGCALIAI NGTIVAQGSQ FSLDDVEVVT ATVDLEEVRS YRAAVMSRGL QASLAEIKFK 300
301 RIDIPVELAL MTSRFDPTVC PTKVREPFYH SPEEEIALGP ACWMWDYLRR CNGTGFFLPL 360
361 SGGIDSCATA MIVHSMCRLV TDAAQNGNEQ VIKDVRKITR SGDDWIPDSP QDLASKIFHS 420
421 CFMGTENSSK ETRNRAKDLS NAIGSYHVDL KMDSLVSSVV SLFEVATGKK PIYKIFGGSQ 480
481 IENLALQNIQ ARLRMVLSYL FAQLLPWVRG IPNSGGLLVL GSANVDECLR GYLTKYDCSS 540
541 ADINPIGGIS KTDLKRFIAY ASKQYNMPIL NDFLNATPTA ELEPMTKDYV QSDEIDMGMT 600
601 YEELGVFGYL RKVEKCGPYS MFLKLLHQWS PKLTPRQISE KVKRFFFFYA INRHKQTVLT 660
661 PSYHAEQYSP EDNRFDLRPF LINPRFPWAS RKIDEVVEQC EAHKGSTLDI MSID |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CPR6 QNS1 |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Ho Y, et al. (2002) |
|
ADH2 CAF120 CPR6 QNS1 RCN2 SLP1 TRR1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
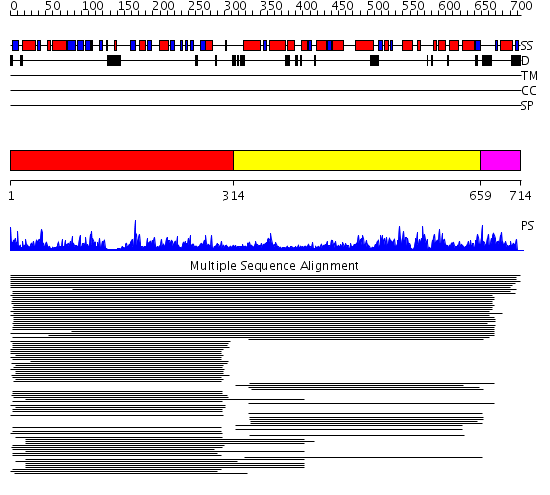
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..313] | 51.39794 | hypothetical protein yl85 | |
| 2 | View Details | [314..658] | 133.27013 | NH3-dependent NAD+-synthetase | |
| 3 | View Details | [659..714] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)