













| General Information: |
|
| Name(s) found: |
ELP2 /
YGR200C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 788 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA transcription elongation factor complex [IDA cytoplasm [IDA |
| Biological Process: |
protein urmylation
[IMP tRNA modification [IMP regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVECITPEAI FIGANKQTQV SDIHKVKKIV AFGAGKTIAL WDPIEPNNKG VYATLKGHEA 60
61 EVTCVRFVPD SDFMVSASED HHVKIWKFTD YSHLQCIQTI QHYSKTIVAL SALPSLISVG 120
121 CADGTISIWR QNIQNDEFGL AHEFTIKKGF FYPLCLSLSK VEEKKYLLAI GGTNVNVFIA 180
181 SFILSDSGIE KCRVVAELEG HEDWVKSLAF RHQETPGDYL LCSGSQDRYI RLWRIRINDL 240
241 IDDSEEDSKK LTLLSNKQYK FQIDDELRVG INFEALIMGH DDWISSLQWH ESRLQLLAAT 300
301 ADTSLMVWEP DETSGIWVCS LRLGEMSSKG ASTATGSSGG FWSCLWFTHE RMDFFLTNGK 360
361 TGSWRMWATK DNIICDQRLG ISGATKDVTD IAWSPSGEYL LATSLDQTTR LFAPWIYDAS 420
421 GRKREIATWH EFSRPQIHGY DMICVETVTD TRFVSGGDEK ILRSFDLPKG VAGMLQKFVG 480
481 IQFEEKSEMP DSATVPVLGL SNKAGEDDAN EDDEEEEGGN KETPDITDPL SLLECPPMED 540
541 QLQRHLLWPE VEKLYGHGFE ITCLDISPDQ KLIASACRSN NVQNAVIRIF STENWLEIKP 600
601 ALPFHSLTIT RLKFSKDGKF LLSVCRDRKW ALWERNMEDN TFELRFKNEK PHTRIIWDAD 660
661 WAPLEFGNVF VTASRDKTVK VWRHQKEPAD DYVLEASIKH TKAVTAISIH DSMIREKILI 720
721 SVGLENGEIY LYSYTLGKFE LITQLNEDIT PADKITRLRW SHLKRNGKLF LGVGSSDLST 780
781 RIYSLAYE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ELP2 ELP3 ELP4 ELP6 IKI1 IKI3 YIL096C |
| View Details | Krogan NJ, et al. (2006) |
|
ELP2 ELP3 ELP4 IKI1 IKI3 KTI12 YIL096C |
| View Details | Gavin AC, et al. (2006) |

|
ELP2 ELP3 ELP4 IKI1 IKI3 |
| View Details | Gavin AC, et al. (2002) |
|
ELP2 ELP3 ELP4 IKI1 IKI3 |
| View Details | Ho Y, et al. (2002) |
|
ELP2 ELP3 IKI3 NIS1 RSF2 |
| View Details | Qiu et al. (2008) |
|
CHD1 ELP2 ELP3 ELP4 ELP6 IKI1 IKI3 RTF1 SPT16 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
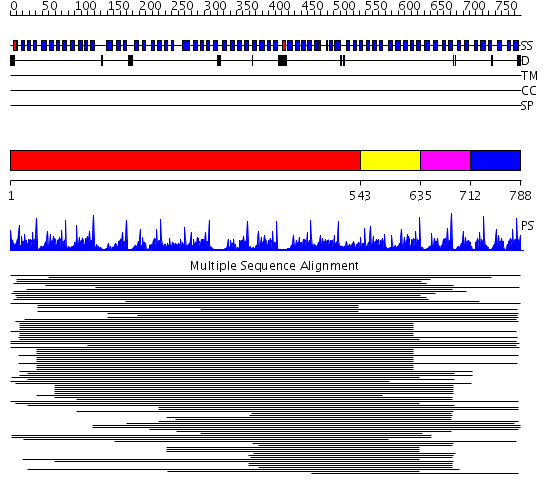
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..542] | 6.522879 | Tricorn protease; Tricorn protease N-terminal domain; Tricorn protease domain 2 | |
| 2 | View Details | [543..634] | 90.927757 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 3 | View Details | [635..711] | 136.9897 | Tup1, C-terminal domain | |
| 4 | View Details | [712..788] | 1.456995 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)