













| General Information: |
|
| Name(s) found: |
TRR1 /
YDR353W
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 319 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[TAS |
| Biological Process: |
response to oxidative stress
[IGI |
| Molecular Function: |
thioredoxin-disulfide reductase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVHNKVTIIG SGPAAHTAAI YLARAEIKPI LYEGMMANGI AAGGQLTTTT EIENFPGFPD 60
61 GLTGSELMDR MREQSTKFGT EIITETVSKV DLSSKPFKLW TEFNEDAEPV TTDAIILATG 120
121 ASAKRMHLPG EETYWQKGIS ACAVCDGAVP IFRNKPLAVI GGGDSACEEA QFLTKYGSKV 180
181 FMLVRKDHLR ASTIMQKRAE KNEKIEILYN TVALEAKGDG KLLNALRIKN TKKNEETDLP 240
241 VSGLFYAIGH TPATKIVAGQ VDTDEAGYIK TVPGSSLTSV PGFFAAGDVQ DSKYRQAITS 300
301 AGSGCMAALD AEKYLTSLE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
TRR1 TRR2 |
| View Details | Krogan NJ, et al. (2006) |

|
TRR1 TRR2 |
| View Details | Ho Y, et al. (2002) |
|
AAT2 ACS2 ADE3 ADE6 ADH4 ADO1 ALA1 APE2 ARA1 ARG4 BAT1 CAF120 CAR2 CDC10 CDC60 CLU1 CPR1 CPR6 CYS3 DED81 DPS1 ENP1 ERG10 ERG20 GCY1 GPH1 GRS1 GSP2 HOM2 HYP2 ILS1 IMD3 KRS1 LSP1 MDH1 MDH3 MPC54 NAS6 OLA1 OYE2 PAB1 PDC5 PFK2 PGM2 PMI40 PRP6 QNS1 RHO5 SAC6 SCP160 SOD1 SSZ1 SUI1 THR4 TPS1 TRR1 UBA1 YDL124W YGK3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
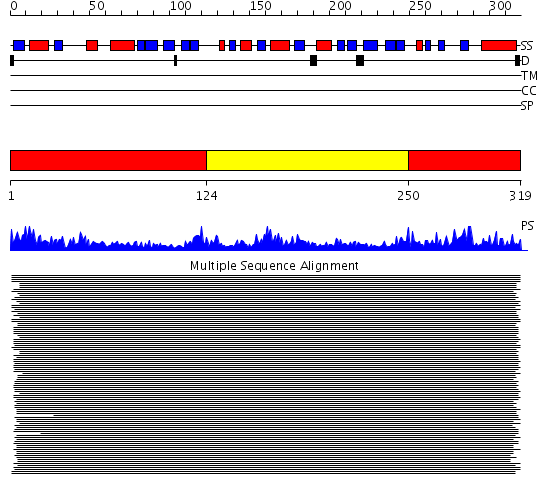
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] [250..319] |
1161.09691 | Thioredoxin reductase | |
| 2 | View Details | [124..249] | 1161.09691 | Thioredoxin reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.56 |
Source: Reynolds et al. (2008)