













| General Information: |
|
| Name(s) found: |
TFB1 /
YDR311W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: |
holo TFIIH complex
[IDA nucleotide-excision repair factor 3 complex [IDA |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter, mitotic
[TAS nucleotide-excision repair, DNA duplex unwinding [TAS nucleotide-excision repair [IDA transcription initiation from RNA polymerase II promoter [TAS |
| Molecular Function: |
general RNA polymerase II transcription factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHSGAAIFE KVSGIIAINE DVSPAELTWR STDGDKVHTV VLSTIDKLQA TPASSEKMML 60
61 RLIGKVDESK KRKDNEGNEV VPKPQRHMFS FNNRTVMDNI KMTLQQIISR YKDADIYEEK 120
121 RRREESAQHT ETPMSSSSVT AGTPTPHLDT PQLNNGAPLI NTAKLDDSLS KEKLLTNLKL 180
181 QQSLLKGNKV LMKVFQETVI NAGLPPSEFW STRIPLLRAF ALSTSQKVGP YNVLSTIKPV 240
241 ASSENKVNVN LSREKILNIF ENYPIVKKAY TDNVPKNFKE PEFWARFFSS KLFRKLRGEK 300
301 IMQNDRGDVI IDRYLTLDQE FDRKDDDMLL HPVKKIIDLD GNIQDDPVVR GNRPDFTMQP 360
361 GVDINGNSDG TVDILKGMNR LSEKMIMALK NEYSRTNLQN KSNITNDEED EDNDERNELK 420
421 IDDLNESYKT NYAIIHLKRN AHEKTTDNDA KSSADSIKNA DLKVSNQQML QQLSLVMDNL 480
481 INKLDLNQVV PNNEVSNKIN KRVITAIKIN AKQAKHNNVN SALGSFVDNT SQANELEVKS 540
541 TLPIDLLESC RMLHTTCCEF LKHFYIHFQS GEQKQASTVK KLYNHLKDCI EKLNELFQDV 600
601 LNGDGESMSN TCTAYLKPVL NSITLATHKY DEYFNEYNNN SN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
SSL1 TFB1 TFB3 TFB4 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CCL1 KIN28 RAD2 RAD3 SSL1 SSL2 TFB1 TFB2 TFB3 TFB4 |
| View Details | Krogan NJ, et al. (2006) |
|
CCL1 ESP1 KIN28 RAD3 SSL1 TFB1 TFB3 TFB4 |
| View Details | Gavin AC, et al. (2006) |
|
SSL1 TFB1 TFB4 |
| View Details | Gavin AC, et al. (2002) |
|
CCL1 DCS2 DNA2 ECM29 FAP1 FPR1 KAP95 KIN28 MEC1 MPH1 MRPS28 MSH2 MSH6 PGK1 RAD23 RAD34 RAD4 RAD52 RFA1 RFA2 RPN10 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT6 SPT7 SRO7 SSL1 TFA1 TFB1 TFB3 TFB4 TIF6 |
| View Details | Qiu et al. (2008) |
|
ELC1 KIN28 RAD3 SSL1 SSL2 TAF2 TFA1 TFA2 TFB1 TFB2 TFB3 TFB4 TFB5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
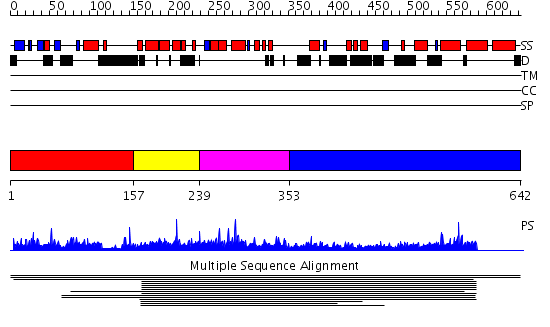
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [157..238] | 11.60206 | BSD domain No confident structure predictions are available. | |
| 3 | View Details | [239..352] | 15.69897 | BSD domain No confident structure predictions are available. | |
| 4 | View Details | [353..642] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [570..642] | 2.034993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)