













| General Information: |
|
| Name(s) found: |
EFB1 /
YAL003W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 206 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ribosome
[TAS eukaryotic translation elongation factor 1 complex [IMP |
| Biological Process: |
translational elongation
[TAS |
| Molecular Function: |
translation elongation factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASTDFSKIE TLKQLNASLA DKSYIEGTAV SQADVTVFKA FQSAYPEFSR WFNHIASKAD 60
61 EFDSFPAASA AAAEEEEDDD VDLFGSDDEE ADAEAEKLKA ERIAAYNAKK AAKPAKPAAK 120
121 SIVTLDVKPW DDETNLEEMV ANVKAIEMEG LTWGAHQFIP IGFGIKKLQI NCVVEDDKVS 180
181 LDDLQQSIEE DEDHVQSTDI AAMQKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
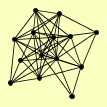
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ABP1 CAM1 CPR1 EFB1 GUS1 HSP104 HXK1 HYP2 RPL40B, RPL40A SSB2 SSZ1 TEF4 YEF3 ZUO1 |
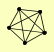
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CAM1 EFB1 HBS1 TEF1, TEF2 TEF4 |
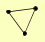
| View Details | Gavin AC, et al. (2006) |
|
CAM1 EFB1 TEF1, TEF2 |
| View Details | Gavin AC, et al. (2002) |
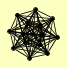
|
CAM1 EFB1 FAP1 GFA1 GUS1 ILV1 KAP123 MGA2 NEW1 OLA1 SBP1 SEC27 TEF1, TEF2 TEF4 |
| View Details | Ho Y, et al. (2002) |
|
ADK1 CDC33 DUG2 EFB1 SEC53 TEM1 VMA5 YHB1 |
| View Details | Qiu et al. (2008) |
|
EFB1 TEF1, TEF2 TEF4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample 3928 - Asf1-S-TEV-ZZ (in hir3delta mutant) | Green EM, et al (2005) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
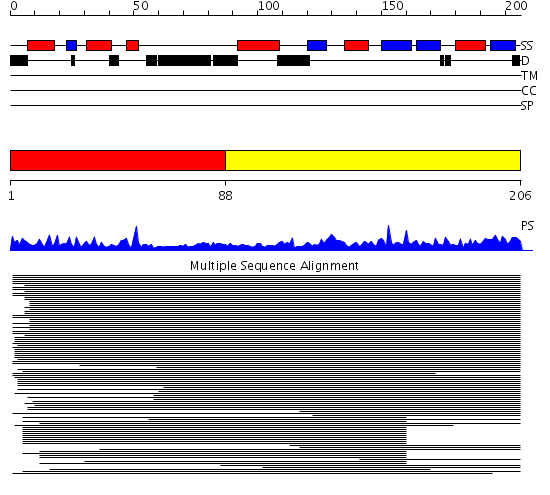
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 5.69897 | Class delta GST | |
| 2 | View Details | [88..206] | 464.741366 | Guanine nucleotide exchange factor (GEF) domain from elongation factor-1 beta |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)